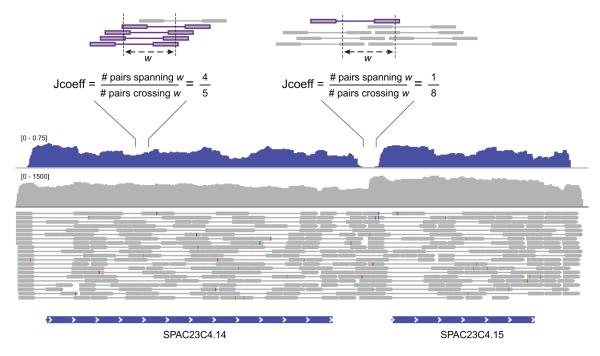
Figure 8. Evaluating paired-read support via the Jaccard similarity coefficient.
Read pair support is computed by first counting the number of RNA-Seq fragments (bounds of paired reads) that span each of two outer points of a specified window length (default: 100 bases), and then computing the Jaccard similarity coefficient (intersection/union) comparing the fragments that overlap either point. An example is shown for a neighboring pair of S. pombe transcripts (SPAC23C4.14 and SPAC23C4.15, bottom) that have substantial overlapping read coverage (gray track), resulting in a contiguous (fused) transcript assembled by Inchworm. However, the Jaccard similarity coefficient (blue track) calculated from the paired-reads (grey dumbbells) clearly identifies the position of reduced pair support. Examples of strong (upper left) and weak (upper right) pair support are depicted at top. When using the ‘--jaccard_clip’ parameter, the Inchworm contig is dissected to two separate full-length transcripts, which are then further processed by Chrysalis and Butterfly as part of the Trinity pipeline.