Abstract
We have shown that the uniaxial cyclic tensile strain of magnitude 10% promotes and enhances osteogenesis of human mesenchymal stem cells (hMSC) and human adipose-derived stem cells (hASC) from normal, nonosteoporotic donors. In the present study, MSC from osteoporotic donors were analyzed for changes in mRNA expression in response to 10% uniaxial tensile strain to identify potential mechanisms underlying the use of this mechanical loading paradigm for prevention and treatment of osteoporosis. Human MSC isolated from three female, postmenopausal osteoporotic donors were analyzed for their responses to mechanical loading using microarray analysis of over 47,000 gene probes. Human MSC were seeded in three-dimensional collagen type I constructs to mimic the organic extracellular matrix of bone and 10% uniaxial cyclic tensile strain was applied to promote osteogenesis. Seventy-nine genes were shown to be regulated within hMSC from osteoporotic donors in response to 10% cyclic tensile strain. Upregulation of six genes were further confirmed with real-time RT-PCR: jun D proto-oncogene (JUND) and plasminogen activator, urokinase receptor (PLAUR), two genes identified as potential key molecules from network analysis; phosphoinositide-3-kinase, catalytic, delta polypeptide (PIK3CD) and wingless-type MMTV integration site family, member 5B (WNT5B), two genes with known importance in bone biology; and, PDZ and LIM domain 4 (PDLIM4) and vascular endothelial growth factor A (VEGFA), two genes that we have previously shown are significantly regulated in hASC in response to this mechanical stimulus. Function analysis indicated that 10% cyclic tensile strain induced expression of genes associated with cell movement, cell proliferation, and tissue development, including development in musculoskeletal and cardiovascular systems. Our results demonstrate that hMSC from aged, osteoporotic donors are capable of enhanced osteogenic differentiation in response to 10% cyclic tensile strain with significant increases in the expression of genes associated with enhanced cell proliferation, musculoskeletal development, and angiogenesis. Surprisingly, cyclic tensile strain of magnitude 10% not only enhanced osteogenesis in hMSC from osteoporotic donors, but also enhanced expression of angiogenic factors. Better understanding and methodologies to promote osteogenesis in hMSC from elderly, osteoporotic donors may greatly facilitate achieving long-term success in bone regeneration and functional bone tissue engineering for this ever-growing patient population.
Introduction
Osteoporosis results in progressive bone loss and an increased risk of bone fracture. Although typical treatment involves the use of pharmacological agents to inhibit bone resorption or increase bone mineral density,1 pharmacotherapy alone is not sufficient for the comprehensive management of osteoporosis. Mechanical loading is also critical for the maintenance of bone homeostasis.2 Regular exercise has been shown to have a significant beneficial effect in the treatment of osteoporosis.3
In general, mechanical loading regulates bone homeostasis through its direct effects on bone formation and remodeling. Bone responds to weight bearing by mineralization and to immobilization by demineralization.4,5 Immobilization in plaster, bed rest, and/or weightlessness all result in calcium loss and lead to disuse osteoporosis.4 However, actively loaded bone exhibits increases in overall bone strength with associated remodeling and increased bone mass.6,7
The dominant mechanical stimuli in bone include fluid shear stress, compression, and tensile strain. Tensile strain plays a key role in new bone regeneration during distraction osteogenesis (DO), an induced bone formation process commonly performed for bone lengthening and the correction of craniofacial deformities. We have previously shown in a mechanobiological study of mandibular DO that tensile stains at magnitudes of 10–12.5% stimulate in vivo bone formation.8,9 Cyclic tensile strain of these magnitudes have also been found to promote cell proliferation and upregulation of bone marker genes in mesenchymal stem cells (MSC), osteoblasts, and periosteal cells.10–12 Previous studies in our laboratory with human MSC (hMSC) and human adipose-derived stem cells (hASC) have further confirmed that 10% uniaxial cyclic tensile strain enhances osteogenesis of these stem cells by increasing bone markers and cell-mediated calcium accretion.13–15 It is now accepted that proper mechanical stimulation can induce proliferation and differentiation of bone precursor cells resulting in an increase in bone formation and bone mass.16
MSC are precursor cells for bone formation and their lineage specification is directly impacted by the mechanical forces to which they are exposed.17,18 We have previously shown that during osteogenic differentiation of hASC, 10% uniaxial cyclic tensile strain increases osteogenesis and causes upregulation of proinflammatory cytokine regulators and angiogenic factors.19 In this study, MSC isolated from aged, postmenopausal osteoporotic donors were cultured in three-dimensional (3D) collagen constructs and analyzed for changes in mRNA expression in response to 10% uniaxial cyclic tensile strain to attempt to identify potential mechanisms underlying the use of appropriate mechanical loading for prevention and treatment of osteoporosis.
We hypothesized that 10% cyclic tensile strain would enhance osteogenic differentiation of hMSC from osteoporotic donors. We further hypothesized that the results of our microarray analysis would indicate that this process occurred through some of the same signaling pathways found to be regulated during the process of bone formation and also through other distinct signaling molecules such as PDZ and LIM domain 4 (PDLIM4), jun D proto-oncogene (JUND), and vascular endothelial growth factor A (VEGFA) that have not been previously studied in the context of bone formation, but which we have shown regulate hASC osteogenesis during 10% cyclic tensile strain.19
Materials and Methods
Cell isolation, culture, and characterization
Excess human bone fragments were obtained from three age-matched female postmenopausal osteoporotic donors undergoing elective orthopedic surgery (two 78-year-old and one 95-year-old Caucasian female donors) in accordance with an approved IRB protocol at University of North Carolina (UNC)-Chapel Hill (IRB 04:1622). Human MSC were isolated from the tissue using a method based on enzymatic digestion and substrate adherence, as previously described.20 Briefly, bone fragments were washed thoroughly with phosphate-buffered saline (PBS) containing 1% penicillin/streptomycin to remove all nonadherent hematopoietic cells. The fragments were diced into small pieces, then digested in a collagenase XI solution (3 mg/mL Collagenase Type XI-S in an α-modified essential medium [αMEM]) at 37°C for 3 h. Cells were then filtered through a 100-μm cell strainer, centrifuged at 500 g for 5 min, and plated in T-75 flasks in a complete growth medium: αMEM containing 10% fetal bovine serum (FBS; Atlanta Biologicals; lot selected), 2 mM L-glutamine, 100 units/mL penicillin, 100 μg/mL streptomycin. After 24 h, the nonadherent cells were washed out. Human MSC were then characterized by their ability to differentiate down osteogenic and adipogenic pathways as previously described.20,21 All cell culture chemicals and supplies were purchased from Mediatech, Inc. and Gibco BRL unless otherwise noted.
Osteogenic differentiation and calcium profile
To confirm the osteogenic differentiation ability of hMSC from osteoporotic donors, cells were cultured in static monolayer in 12-well plates with either the nondifferentiating growth medium or the osteogenic medium for 14 days. The osteogenic medium consisted of the complete growth medium plus osteogenic growth supplements comprised 50 μM ascorbic acid, 0.1 μM dexamethasone, and 10 mM β-glycerophosphate. After 14 days, osteogenic differentiation was determined by the endogenous alkaline phosphatase (EALP) activity and the deposition of a mineralized matrix.
For the EALP activity, cell monolayers were washed twice with PBS and fixed using 10% formalin for 15 min. The cell membranes were then permeabilized by incubating in 0.2% Triton X-100/0.5% bovine serum albumin in PBS for 10 min. Subsequently, cell monolayers were rinsed again in PBS and EALP was detected by incubating in 20 μL of 20-fold diluted alkaline phosphatase substrate (ELF 97; Molecular Probes) for 15 min. The phosphatase reaction was subsequently stopped by adding 5 mM Levamisol and 25 mM EDTA (Sigma) in PBS (wash buffer).
Deposition of the mineralized matrix was visualized with Alizarin Red staining. In brief, cell monolayers were rinsed with PBS, fixed with 10% formalin for 30 min, rinsed twice in PBS, and stained with 2% Alizarin Red S.
Cell-mediated calcium accretion of hMSC from the three osteoporotic donors was evaluated on day 14 and compared with cell-mediated calcium accretion by hMSC from two young, nonosteoporotic female donors, ages 19 and 25. Cell monolayers were rinsed twice with PBS, then scraped out in 0.5 N HCl. Calcium was dissolved overnight at 4°C and the supernatant analyzed with a colorimetric Calcium LiquiColor® assay (Stanbio Laboratory). Calcium accretion was then normalized to cellular protein quantity (micro BCA protein assay; Pierce). The proliferation rate, based on the metabolic activity of the hMSC, was assessed using the Alamar Blue assay (AbD Serotec) at days 1, 3, 6, and 9.
Fabrication of collagen gels
Human MSC were seeded into 3D collagen gels consisting of 70% type I collagen (BD Biosciences) (pH adjusted to 7.0), 20% 5× MEM, and 10% FBS at 60,000 cells/200 μL gel solution. Linear 3D collagen constructs were created by loading the cell-seeded gel solutions into Tissue Train® collagen I-coated six-well culture plates (Flexcell International). Constructs were allowed to polymerize for 2 h before addition of the complete growth medium.
Application of cyclic tensile strain
Twenty-four hours after cell seeding in the collagen gels, cell-seeded constructs were cultured for an additional 2 weeks in growth media in the presence (experimental) and/or absence (control) of 10% uniaxial cyclic tensile strain. Cell-seeded constructs were subjected to 14 days of 10% uniaxial tensile strain at 1 Hz for 4 h/day using a computer-driven strain device (FX-4000T; Flexcell International). Constructs were collected on day 14 of treatment for RT-PCR and microarray analyses. Constructs were washed twice in PBS, removed from their anchors, placed in a lysis buffer containing β-mercaptoethanol, and frozen at −80°C until RNA could be isolated.
RNA isolation and real-time RT-PCR analysis
Constructs were thawed on ice, ground with a mini-pestle, and homogenized using a QIAshedder (Qiagen). Total RNA was then purified using Eppendorf Perfect RNA mini-columns according to the manufacturer's recommended protocol for eukaryotic cells. For microarray analysis, total RNA was quantitated using a microplate-based RiboGreen® method (Molecular Probes).
Quantification of total RNA for PCR analysis was performed with a Nanodrop system (Thermo-Fisher Scientific). One hundred nanograms of each RNA sample was reverse transcribed into cDNA using SuperScript™ III (Invitrogen) with oligo dT primers. Real-time PCR was performed using an ABI Prism® 7000 Sequence Detection System (Applied Biosystems). Based on findings from the microarray analyses, six genes were selected for further confirmation of expression using real-time RT-PCR. Two genes were selected from potential key molecules identified through network analysis, two genes were selected based on the canonical pathway and their potential role in bone biology, and the final two genes were selected based on their similar expression in a previous study in hASC.19 TaqMan-based PCR Assays-on-Demand™ (Applied Biosystems) were used for further confirmation of mRNA expression of JUND, plasminogen activator, urokinase receptor (PLAUR), wingless-type MMTV integration site family, member 5B (WNT5B), phosphoinositide-3-kinase, catalytic, delta polypeptide (PIK3CD), VEGFA, PDLIM 4, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), the endogenous control.
Expression levels were determined using the ΔΔCT method,22 and presented as a fold change from the mRNA level in normal human adult bone tissue (BioChain), which was set to 1.0. Real-time RT-PCR analyses were performed for n=3 donors with triplicate samples per condition. Data were subjected to a two-tailed Student's t-test to determine significant difference (p<0.05) from control (unloaded, 0% tensile strain). Data are presented as mean±standard error.
Biotin labeling, streptavidin antibody staining, scanning, and detection
Gene expression analysis was conducted using Affymetrix Human Genome U133 2.0 Genechip® arrays (Affymetrix). Total RNA was amplified using the Affymetrix 2-Cycle cDNA Synthesis protocol. Starting with 20 ng of total RNA, cRNA was produced according to the manufacturer's protocol. For each array, 15 μg of amplified cRNAs were fragmented and hybridized to the array for 16 h in a rotating hybridization oven using the Affymetrix Eukaryotic Target Hybridization Controls and protocol. Slides were stained and washed as indicated in the Antibody Amplification Stain for Eukaryotic Target protocol using the Affymetrix Fluidics Station FS450. Arrays were then scanned with an Affymetrix Scanner 3000. Data were obtained using the Genechip Operating Software (Version 1.2.0.037). Microarray analyses were performed for n=3 donors (one sample per condition) on day 14.
Microarray data analysis
Data preprocessing, normalization, and error modeling were performed with the Rosetta Resolver system (version 7.2.) (Rosetta Biosoftware). Principal Component Analysis (PCA) was performed on all samples and all probes to characterize any variability present in the data. Intensity profiles were combined by weighted-averaging into Intensity Experiments. When required, Intensity Experiments were built into ratios representing treated/control (10% uniaxial cyclic tensile strain compared to unloaded, 0% tensile strain), as described by Stoughton and Dai.23 An error-weighted ANOVA with the Bonferroni test was used to reduce the number of false positives with p<0.01.
Whole genome expression data were visualized in the context of molecular function, canonical pathways, and biological network using the Ingenuity Pathway Analysis (IPA) system (version 8.0) (Ingenuity Systems; www.ingenuity.com). Data sets containing gene identifiers and corresponding expression values were uploaded into the application. Each gene identifier was mapped to its corresponding gene. In the case of genes with multi-identifiers, the highest expression value was selected. The function and pathway analysis of IPA was generated based on all publicly available, published knowledge to identify potential regulatory pathways and molecules.
Results
Proliferation and osteogenic differentiation of hMSC from postmenopausal Caucasian osteoporotic donors
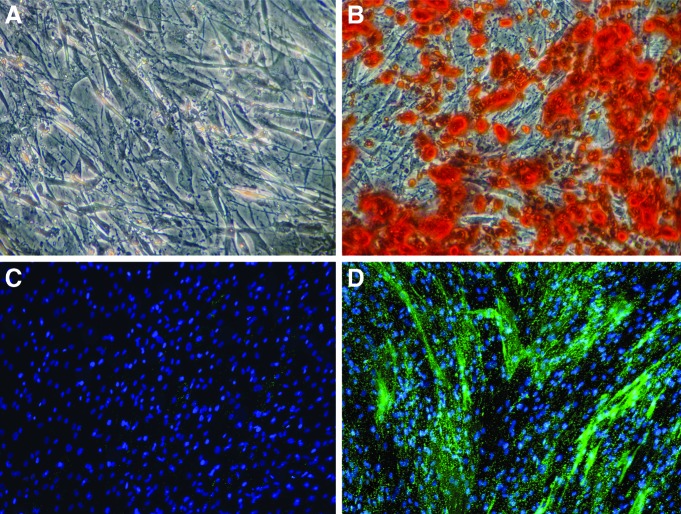
Human MSC from three aged, postmenopausal female osteoporotic donors had been preselected for positive MSC markers and capability for adipogenic and osteogenic differentiation in 2D culture. All three selected cell lines were verified to deposit mineral over an area spanning at least 50% of the tissue culture well, determined by Alizarin Red staining (Fig. 1A, B) and expression of alkaline phosphatase (Fig. 1C, D).
FIG. 1.
Human mesenchymal stem cells (hMSC) from three aged, postmenopausal female osteoporotic donors are able to osteogenically differentiate after chemical stimulation in osteogenic induction media for 14 days in 2D monolayer culture. Alizarin Red staining for calcium accretion (A) in growth media or (B) osteogenic media. Endogenous alkaline phosphatase (EALP) activity of hMSC after 14 days (C) in growth media or (D) in osteogenic induction media. EALP activity indicated in bright green; nuclei shown in blue. Color images available online at www.liebertpub.com/tea
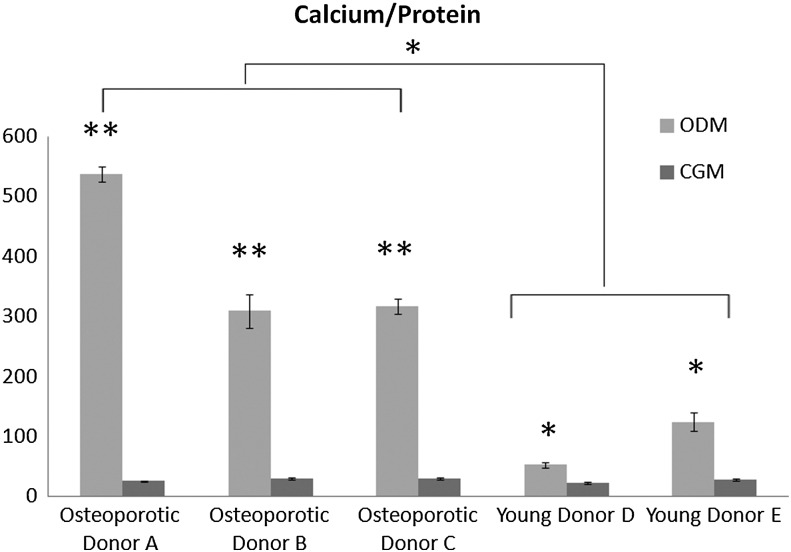
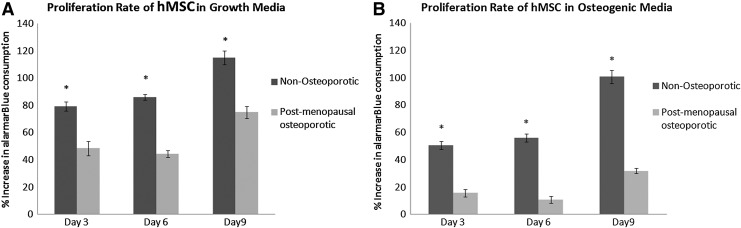
To further evaluate osteogenic differentiation potential, accreted calcium, protein content, and proliferation rate were quantified and compared with hMSC from young, nonosteoporotic donors. Relative to hMSC cultured in complete growth media, calcium content was significantly (p<0.01) increased by osteogenic media in all three osteoporotic donors (Fig. 2). Interestingly, hMSC from the three osteoporotic donors exhibited a significantly (p<0.05) higher calcium/protein ratio than hMSC from young, nonosteoporotic donors. However, the metabolic activity as determined by alamar blue assay on days 3, 6, 9 showed that hMSC from osteoporotic donors proliferated significantly (p<0.05) less than hMSC from young donors during culture in both growth media and osteogenic media in monolayer culture (Fig. 3). These findings indicated that the ability of hMSC from osteoporotic donors to be chemically induced to differentiate down an osteogenic lineage is not less than hMSC from young, nonosteoporotic donors. However, their proliferation rate is diminished.
FIG. 2.
Quantitative analysis of cell-mediated calcium accretion normalized to cellular protein content after 14 days of hMSC culture in the complete growth medium (CGM) or the osteogenic medium (ODM). hMSC isolated from osteoporotic donors (A, B, C) exhibited a significantly (*p<0.05, **p<0.01) higher calcium/protein ratio than hMSC isolated from young, nonosteoporotic donors (D, E).
FIG. 3.
Comparative analysis of average proliferation rate (normalized to day 1) of hMSC isolated from osteopotic donors (OP) and hMSC isolated from young, nonosteoporotic donors (Y) in the complete growth medium (A) or the osteogenic medium (B) on days 3, 6, and 9 in 2D monolayer culture. Proliferation of hMSC from osteoporotic donors is significantly (*p<0.05) less than hMSC from nonosteoporotic donors at all time points in both growth and osteogenic differentiation media.
Function analysis of genes regulated by hMSC in response to 10% uniaxial cyclic tensile strain
It has been shown that exercises associated with increased bone loading increase the bone mineral content in postmenopausal osteoporotic women over the age of 7024 and that mechanical loading increases both hMSC proliferation and osteogenic differentiation.10,16,25–27 Therefore, we further investigated how mechanical loading in the form of 10% uniaxial cyclic tensile strain would regulate expression of genes involved with proliferation and/or osteogenic differentiation of hMSC from osteoporotic donors when hMSC were seeded in 3D collagen I culture. A full microarray analysis with 47,000 transcripts was implemented.
To identify the effect of 10% cyclic tensile strain on hMSC from osteoporotic donors, the gene expression profiles of hMSC cultured in complete growth media in the presence of strain were compared to gene expression profiles of hMSC cultured in growth media in the absence of mechanical load (0% strain). Seventy three transcripts were modulated by strain, which mapped to 68 genes with IPA (Supplementary Table S1 Supplementary Data are available online at www.liebertpub.com/tea). Sixty-two genes were eligible for network analysis and 57 genes were eligible for function analyses.
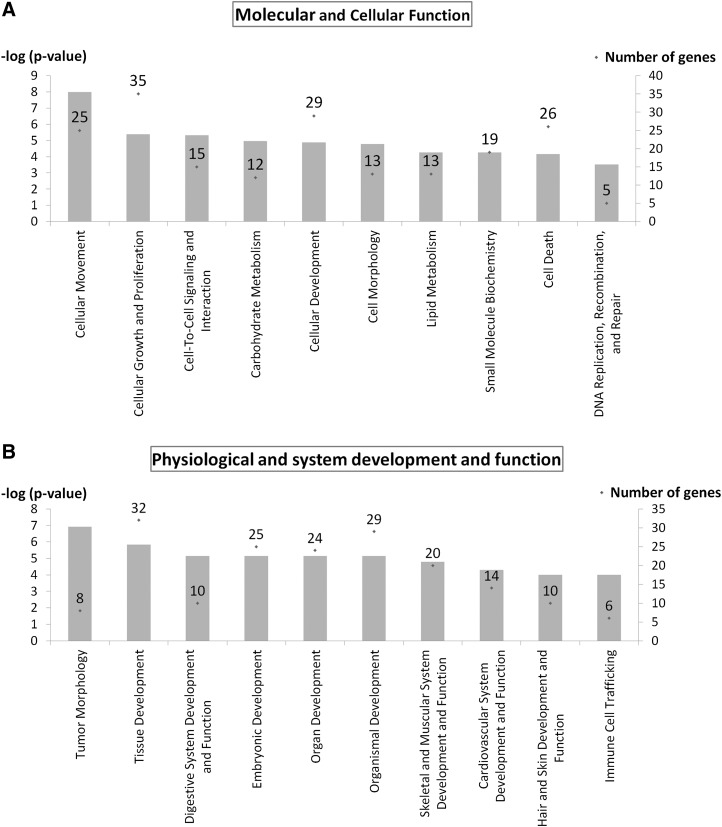
Cellular movement was the top molecular and cellular function when categorizing the genes modulated by strain (25 genes, p<10−8–10−3). Major subfunctions included invasion of cells, cell movement, chemotaxis, and cell migration with upregulation of 23 genes, including transcription regulators JUND, basic helix-loop-helix family, member e40 (BHLHE40), H2.0-like homeobox (HLX), plasma membrane receptor PLAUR, insulin-like growth factor 1 receptor (IGF1R), actin binding FXYD domain containing ion transport regulator 5 (FXYD5), and ezrin (EZR) (Fig. 4A and Supplementary Table S2).
FIG. 4.
Function analysis indicates that 10% uniaxial cyclic tensile strain induces expression of genes associated with cellular movement, growth, and proliferation in hMSC from osteoporotic donors as shown in the top ten molecular and cellular functions (A); and the expression of genes associated with system development and function for skeletal, muscular, and cardiovascular systems as shown in the top ten categories expressed within physiological and system development and function (B). Left Y-axis is −log (p-value) shown as bar chart. Right Y-axis is number of genes associated with functions shown as dot (.) with number.
Tumor morphology was ranked first when categorizing the genes modulated by strain in physiological system development and function (8 genes, p<10−7–10−3) with major subfunctions of tumor progression and vascularization (Fig. 4B and Supplementary Table S3). The second ranked physiological system development and function was tissue development with 32 differentially expressed genes involved (p<10−6–10−3) with the subfunctions of tissue formation, adhesion, development, morphology, and angiogenesis in multiple tissues. Three specific physiological systems also ranked in the top ten of development and function that were differentially expressed by hMSC in response to strain were (1) digestive system (10 genes, p<10−6–10−3) with genes involved with morphology and development of tooth, (2) skeletal and muscular system (20 genes, p<10−5–10−3) with genes involved with development and formation of muscle and bone, and (3) cardiovascular system (14 genes, p<10−5–10−3) with genes involved with angiogenesis, neovascularization, and development of blood vessels (Fig. 4B and Supplementary Table S3).
Network analysis and canonical pathway of genes regulated by hMSC in response to 10% uniaxial cyclic tensile strain
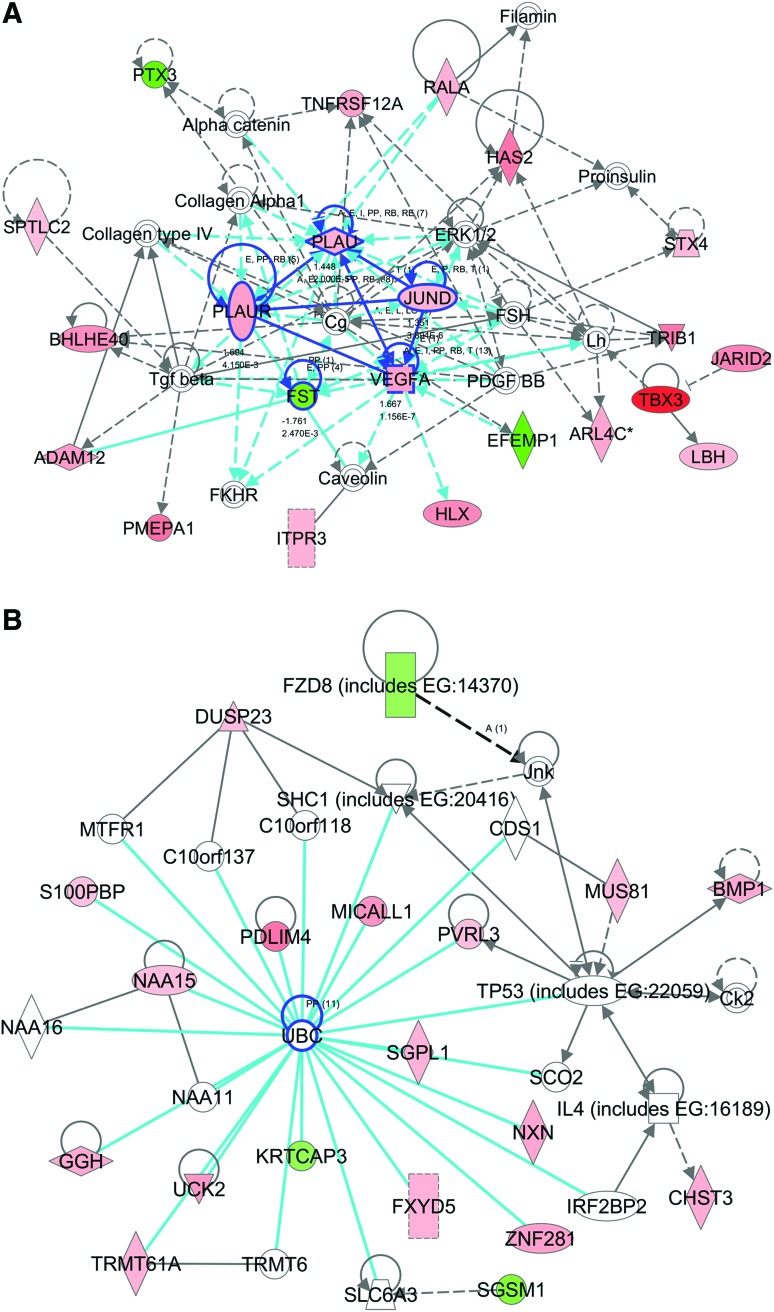
The first ranked network was associated with cellular movement, cell morphology, and skeletal and muscular system development and function. This network had a score of 52 (scores of 2 or higher have at least a 99% confidence of not being generated by random chance alone) with 22 focused genes and 37 total genes in the network (Fig. 5A and Supplementary Table S4). Five focused molecules, JUND, tumor necrosis factor receptor superfamily, member 12A (TNFRSF12A), follistatin (FST), PLAU, and PLAUR, were located in the center of the network and linked with four or more other molecules. The second ranked network was associated with cardiovascular disease, tissue morphology, and molecular transport with a score of 42 and 19 focused genes. In contrast to the first ranked network, centered genes in this network were not expressed by hMSC in response to strain at day 14 (Fig. 5B). The biggest group of genes was directly linked with ubiquitin C (UBC), including upregulation of PDLIM4, zinc finger protein 281 (ZNF281), uridine-cytidine kinase 2 (UCK2), sphinganine-1-phosphate aldolase (SGPL1), FXYD domain containing ion transport regulator 5 (FXYD 5), and MICAL-like 1 (MICALL1).
FIG. 5.
Potential key molecules affected in hMSC from osteoporotic donors in response to 10% uniaxial cyclic tensile strain are jun D proto-oncogene (JUND), plasminogen activator, urokinase (PLAU), plasminogen activator, urokinase receptor (PLAUR), vascular endothelial growth factor A (VEGFA), and follistatin (FST) as centered in the first (A), and ubiquitin C (UBC) as centered in the second (B) ranked networks (highlighted with blue circles). The first ranked network was generated with 22 focus genes previously found to be associated to cellular movement, cell morphology, and skeletal and muscular system development and function (A). The second ranked network was generated with 19 focus genes previously found to be associated with tissue morphology, cardiovascular disease, and molecular transport (B). Red indicates upregulated molecules; green indicates downregulated molecules. Color intensity indicates the level of expression (see Supplementary Table S4 for full names and expression profile data). Color images available online at www.liebertpub.com/tea
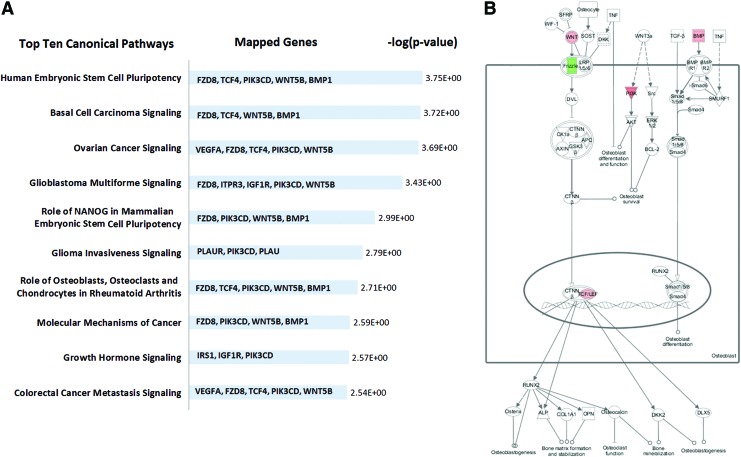
Pathway analysis indicated that relatively few genes (20 of 68 genes) that were modulated in response to strain by hMSC were significantly mapped in the canonical pathways (supplementary Table S5). Interestingly, five of the genes (metalloproteinases BMP1, receptor binding WNT5B, protein binding PIK3CD, transcription factor TCF4, and receptor FZD8) were found to be repeatedly associated and mapped in multiple top ten pathways (Fig. 6).
FIG. 6.
Canonical pathways affected in hMSC from osteoporotic donors by 10% uniaxial cyclic tensile strain. Top 10 canonical pathways showed that five genes (FZD8, TCF4, phosphoinositide-3-kinase, catalytic, delta polypeptide [PIK3CD], wingless-type MMTV integration site family, member 5B [WNT5B], and BMP1) are associated and repeatedly mapped in multiple pathways (A). Mapped canonical pathway, role of osteoblasts, osteoclasts, and chondrocytes in rheumatoid arthritis, showed the integrated signals of these genes in Wnt/BMP/PIK signaling (B). Color images available online at www.liebertpub.com/tea
Real-time RT-PCR analysis of selected differentially expressed genes identified by cDNA microarray
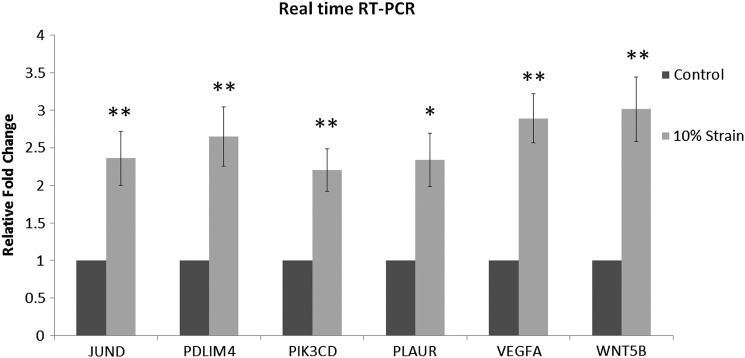
To confirm the expression data from the microarray analyses, six genes upregulated by hMSC in response to strain as detected by microarray were also validated with quantitative RT-PCR analysis. Genes were chosen based on the statistical significance of the differential expression of a gene as detected by microarray (i.e., the error-weighted ANOVA with the Bonferroni test p<0.01) and also their potential functional relevance to bone metabolism and/or potential key molecules expressed by hMSC in response to mechanical loading from pathway, network, and function analyses. Two of the selected genes were centered in the first rank network: JUND, a member of activator protein-1 (AP-1), shown to be a controlling molecule in bone formation, osteoblast proliferation, and osteoblast differentiation,28–31 and PLAUR, shown to be involved in bone homeostasis and bone formation.32,33 PIK3CD and WNT5B were both mapped together with the top five canonical pathways (Fig. 6). PIK3CD, a gene encoding for a catalytic subunit of p110, has been found to be upregulated during bone fracture repair34 and to play an important role in osteoblast differentiation.35 PDLIM4 was one of the top ten upregulated genes in our data set and has previously been shown to have polymorphisms found to associate with bone mineral density regulation.36,37 VEGFA is an angiogenic factor. We have previously reported that both VEGFA and PDLIM4 are upregulated by hASC during osteogenesis in response to 10% uniaxial cyclic tensile strain.19 RT-PCR results confirmed that expression of all genes was significantly increased in hMSC in response to 10% uniaxial cyclic tensile strain (Fig. 7).
FIG. 7.
RT-PCR results. Upregulation of six genes identified with microarray analysis: JUND, PDZ and LIM domain 4 (PDLIM 4), PIK3CD, PLAUR, VEGFA, and WNT5B in hMSC from osteoporotic donors in response to 10% uniaxial cyclic tensile strain had expression confirmed with RT-PCR. Gene expression in growth media without the application of strain set to 1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) normalization.
Discussion
Human MSC hold great promise for their use in cell-based therapy for a number of degenerative conditions. Whereas many previous studies have been performed on the use of MSC for tissue engineering and shown their promise for regeneration of multiple tissues,38–43 only a limited amount of research has focused on hMSC from aged donors who more typically suffer from degenerative diseases. In the present study, hMSC from aged, postmenopausal osteoporotic donors were analyzed using microarray analysis to determine potential mechanobiological mechanisms associated with enhanced osteogenic differentiation of hMSC from osteoporotic donors. Collagen type I was used to mimic the organic extracellular matrix (ECM) of bone and 10% uniaxial cyclic tensile strain was applied as an appropriate mechanical load to promote osteogenesis.13–15,19,44 Microarray analysis with over 30,000 gene probes was utilized for global analysis to obtain insight into associated regulatory mechanisms.
We found that hMSC from aged, osteoporotic donors exhibit a significant decrease in the proliferation rate compared to hMSC from young, nonosteoporotic donors. These findings are consistent with previous investigators who have shown that hMSC progenitor cell numbers are reduced with aging.45 However, the hMSC from osteoporotic donors were able to differentiate down an osteogenic pathway when chemically induced with a medium that included osteogenic supplements consisting of ascorbic acid, dexamethasone, and β-glycerophosphate, to a greater extent on a per cell basis than hMSC from young donors. Initially, one might infer that this result could occur through use of isolated hMSC, which were preinduced in vivo with pharmacotherapy for osteoporosis; however, the selected donors chosen for this study had no history of osteoporosis pharmacotherapy treatment. Previous studies on the effects of aging on osteogenic differentiation of hMSC have reported contradictory results depending on grouping and different conditions of the donors.46–51 Therefore, in this study, we specifically compared differentiation of hMSC from postmenopausal osteoporotic donors over the age of 60 who were not undergoing any pharmacotherapy, to those of healthy, skeletally mature females between the ages of 19–25.
One hundred eighty-four transcripts and 68 transcripts that we previously found to be significantly modulated by 10% cyclic tensile strain in hASC19 were also modulated by hMSC from osteoporotic donors in response to this mechanical stimulus. One might expect hMSC to have higher sensitivity to mechanical loading than hASC as MSC are direct progenitor cells in bone, a highly load bearing tissue. However, a combination of the use of hASC from younger donors as well as the additional induction by the osteogenic differentiation medium utilized in the previous study might have provided synergistic effects to further increase the mechanosensitivity of hASC. This should be further explored in future studies.
Eight genes were significantly upregulated in both hASC and hMSC by 10% cyclic tensile strain (Supplementary Table S6). Similarities in mechanobiological responses included upregulation of genes associated with functions such as cell proliferation, cellular movement, and cardiovascular development (Supplementary Fig. S1).
Function analysis showed an increase in genes associated with cellular movement and proliferation of hMSC from osteoporotic donors when exposed to 10% uniaxial cyclic tensile strain in 3D collagen culture. Although relative proliferation of hMSC in 3D collagen gels by cyclic tensile strain was not monitored over time in this study, we observed a higher yield of total RNA content in those hMSC-seeded constructs that underwent mechanical loading (Supplementary Fig. S2). It has been shown that cell proliferation can be promoted with appropriate mechanical loading both in vivo and in vitro.52–55 Our results support these previous findings. Whereas tumor morphology was ranked first for physiology function, only 8 genes were associated with this function. However, 32 genes were associated with the second ranked tissue development function, and 20 genes were associated with the seventh ranked skeletal and muscular system development and function (Fig. 4B). The lower number of mapped genes associated with tumor morphology likely result from the low number of genes associated with this function in the publicly available literature. These same 8 genes were subsets of tissue development and skeletal and muscular system development. Therefore, to better identify the potential function of consistently expressed genes, both the number of mapped genes and the p-value for each function were simultaneously considered for more accurate analysis.
Physiological function showed not only an increase in musculoskeletal development, but also an increase in cardiovascular development by hMSC from osteoporotic donors exposed to 10% cyclic tensile strain. We have previously shown that 10% cyclic tensile strain promotes osteogenic differentiation of hMSC and hASC in both 2D and 3D culture.13–15,19,44 Previous studies with human periodontal ligament stem cells, preosteoblasts, and osteoblasts have also reported that tensile strain induces osteogenesis and ECM formation.56–58 We have also previously found that 10% cyclic tensile strain induces the upregulation of proinflammatory cytokine regulators: SOCS3 and IL1RN, and angiogenic factors: FGF2, VEGFA, and MMP2 in hASC undergoing osteogenic differentiation.19 Findings from the current study indicate that upregulation of VEGF in response to 10% cyclic tensile strain is consistent in both hASC and hMSC. Our finding of enhanced angiogenic signaling by MSC in response to specific mechanical stimulation is consistent with other findings. It has been shown that with cyclic tensile induction, MSC can promote angiogenesis by elevating proliferation of endothelial cells, potentially through FGFR and VEGFR signaling.59 In a recent study with VEGF-deficient mice and mice with VEGF receptor 1 or VEGF receptor 2 knockdown, it was shown that VEGF not only played a role in angiogenesis, but also that intracellular VEGF regulated the balance of osteoblast or adipocyte differentiation in MSC.60 Whereas the focus of the present study was on a complete microarray gene expression analysis of MSC from osteoporotic donors in response to mechanical load in the form of 10% cyclic tensile strain alone, future studies examining the combination of osteogenic induction media and cyclic tensile strain on MSC response would be interesting to perform. Genes shown to be modulated in this study could be further investigated as well as other genes of interest in the context of bone formation or cellular function in response to tensile strain, such as BHLHE40, HLX, IGF1R, FXYD5, and EZR.
Interestingly, downregulation of FZD8 and upregulation of WNT5B, TCF4, PIK3CD, and BMP1 in hMSC with 10% uniaxial cyclic tensile strain were repeatedly associated together and mapped in multiple canonical pathways (Fig. 6). FZD8, WNT5B, and TCF4 are members of the Wnt signaling pathway. WNT5B and FZD8, a Wnt receptor-ligand complex, have also been found to be modulated in different phases of osteogenic differentiation of hMSC when chemically induced in 2D monolayer culture.61 Downregulation of FZD8 and upregulation of its inhibitor have also been shown to respond as a feedback inhibition when Wnt signaling is upregulated in human embryonic stem cells.62 TCF4, one of the nuclear effectors of Wnt signaling, represents a final target of cellular control through Wnt-dependent growth and survival.63 Wnt signaling pathways have been found to be integrated together with BMP signaling and phosphoinositide 3-kinase-dependent signaling in stem cells' self-renewal and osteoblast differentiation.64–66 We have shown here that BMP-1 (from BMP signaling), PIK3CD (from PIKs signaling), and FZD8, WNT5B, and TCF4 (from Wnt signaling) are modulated together in the response of hMSC from osteoporotic donors to 10% uniaxial cyclic tensile strain as a potential integration of Wnt/BMP/PIK signaling. Whereas beyond the scope of this study, further investigation of these signaling pathways in response to 10% cyclic tensile strain should be performed to fully evaluate their independent and integrated signaling mechanisms.
In summary, we have found that 10% cyclic tensile strain enhances the modulation of genes associated with increases in both proliferation and osteogenic differentiation of hMSC from osteoporotic donors. The potential mechanisms include the known canonical pathways that play an important role in bone formation such as Wnt/BMPs/PIKs signaling and, also, interestingly, through the process of angiogenesis and neovascularization. This is the first study to investigate the effect of both 3D collagen culture and 10% uniaxial cyclic tensile strain on osteogenic differentiation of hMSC from osteoporotic donors. A complete microarray analysis of the expression of 47,000 genes with identifying potential key genes and signaling was performed. The findings from this study provide significant information and better understanding toward our use of hMSC from elderly, osteoporotic donors for achieving long-term success in bone regeneration and functional bone tissue engineering for this ever-growing patient population.
Supplementary Material
Acknowledgments
This research was supported, in part, by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, by the National Center for Research. Resources grant 10KR51023 (E.G.L.), by NIH/NIBIB grant R03EB008790 (E.G.L.), and by NSF/CBET grant 1133427 (E.G.L.). Excess human bone fragments were obtained from waste tissue from voluntary orthopedic surgeries at the UNC-CH hospital. Cell isolation, culture, mechanical loading, and PCR analysis were performed at the North Carolina State University. Microarray imaging and statistical analysis were performed at the National Institute of Environmental Health Sciences, NIH.
Disclosure Statement
No competing financial interests exist.
References
- 1.Siris E.S., Selby P.L., Saag K.G., Borgström F., Herings R., and Silverman S.L.Impact of osteoporosis treatment adherence on fracture rates in North America and Europe. Am J Med 122,S3, 2009 [DOI] [PubMed] [Google Scholar]
- 2.Sinaki M., Pfeifer M., Preisinger E., Itoi E., Rizzoli R., Boonen S., Geusens P., and Minne H.W.The role of exercise in the treatment of osteoporosis. Curr Osteoporos Rep 8,138, 2010 [DOI] [PubMed] [Google Scholar]
- 3.Hurley B., and Armstrong T.J.Bisphosphonates vs exercise for the prevention and treatment of osteoporosis. J Nurse Pract 8,217, 2012 [Google Scholar]
- 4.Leblanc A.D., Schneider V.S., Evans H.J., Engelbretson D.A., and Krebs J.M.Bone mineral loss and recovery after 17 weeks of bed rest. J Bone Miner Res 5,843, 1990 [DOI] [PubMed] [Google Scholar]
- 5.Wolff I., Van Croonenborg J., Kemper H., Kostense P., and Twisk J.The effect of exercise training programs on bone mass: a meta-analysis of published controlled trials in pre-and postmenopausal women. Osteoporos Int 9,1, 1999 [DOI] [PubMed] [Google Scholar]
- 6.Turner C.H., and Robling A.G.Designing exercise regimens to increase bone strength. Exerc Sport Sci Rev 31,45, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Robling A.G., Castillo A.B., and Turner C.H.Biomechanical and molecular regulation of bone remodeling. Annu Rev Biomed Eng 8,455, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Loboa E.G., Fang T.D., Parker D.W., Warren S.M., Fong K.D., Longaker M.T., and Carter D.R.Mechanobiology of mandibular distraction osteogenesis: finite element analyses with a rat model. J Orthop Res 23,663, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Loboa E.G., Fang T.D., Warren S.M., Lindsey D.P., Fong K.D., Longaker M.T., and Carter D.R.Mechanobiology of mandibular distraction osteogenesis: experimental analyses with a rat model. Bone 34,336, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Kanno T., Takahashi T., Ariyoshi W., Tsujisawa T., Haga M., and Nishihara T.Tensile mechanical strain up-regulates Runx2 and osteogenic factor expression in human periosteal cells: implications for distraction osteogenesis. J Oral Maxillofac Surg 63,499, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Fong K.D., Nacamuli R.P., Loboa E.G., Henderson J.H., Fang T.D., Song H.M., Cowan C.M., Warren S.M., Carter D.R., and Longaker M.T.Equibiaxial tensile strain affects calvarial osteoblast biology. J Craniofac Surg 14,348, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Koike M., Shimokawa H., Kanno Z., Ohya K., and Soma K.Effects of mechanical strain on proliferation and differentiation of bone marrow stromal cell line ST2. J Bone Miner Metab 23,219, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Hanson A.D., Marvel S.W., Bernacki S.H., Banes A.J., van Aalst J., and Loboa E.G.Osteogenic effects of rest inserted and continuous cyclic tensile strain on hASC lines with disparate osteodifferentiation capabilities. Ann Biomed Eng 37,955, 2009 [DOI] [PubMed] [Google Scholar]
- 14.Sumanasinghe R.D., Bernacki S.H., and Loboa E.G.Osteogenic differentiation of human mesenchymal stem cells in collagen matrices: effect of uniaxial cyclic tensile strain on bone morphogenetic protein (BMP-2) mRNA expression. Tissue Eng 12,3459, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Wall M.E., Rachlin A., Otey C.A., and Loboa E.G.Human adipose-derived adult stem cells upregulate palladin during osteogenesis and in response to cyclic tensile strain. Am J Physiol Cell Physiol 293,2007 [DOI] [PubMed] [Google Scholar]
- 16.Qi M.C., Hu J., Zou S.J., Chen H.Q., Zhou H.X., and Han L.C.Mechanical strain induces osteogenic differentiation: Cbfa1 and Ets-1 expression in stretched rat mesenchymal stem cells. Int J Oral Maxillofac Surg 37,453, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Turner C.H., Forwood M., Rho J.Y., and Yoshikawa T.Mechanical loading thresholds for lamellar and woven bone formation. J Bone Miner Res 9,87, 1994 [DOI] [PubMed] [Google Scholar]
- 18.Kelly D., and Prendergast P.J.Mechano-regulation of stem cell differentiation and tissue regeneration in osteochondral defects. J Biomech 38,1413, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Charoenpanich A., Wall M.E., Tucker C.J., Andrews D.M.K., Lalush D.S., and Loboa E.G.Microarray analysis of human adipose-derived stem cells in three-dimensional collagen culture: osteogenesis inhibits bone morphogenic protein and Wnt signaling pathways, and cyclic tensile strain causes upregulation of proinflammatory cytokine regulators and angiogenic factors. Tissue Eng Part A 17,2615, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakaguchi Y., Sekiya I., Yagishita K., Ichinose S., Shinomiya K., and Muneta T.Suspended cells from trabecular bone by collagenase digestion become virtually identical to mesenchymal stem cells obtained from marrow aspirates. Blood 104,2728, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Wall M.E., Bernacki S.H., and Loboa E.G.Effects of serial passaging on the adipogenic and osteogenic differentiation potential of adipose-derived human mesenchymal stem cells. Tissue Eng 13,1291, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Livak K.J., and Schmittgen T.D.Analysis of relative gene expression data using real-time quantitative PCR and the 2 CT method. Methods 25,402, 2001 [DOI] [PubMed] [Google Scholar]
- 23.Stoughton R., and Dai H.Statistical combining of cell expression profiles, U.S. Patent No 6,351,712, 2002 [Google Scholar]
- 24.Simkin A., Ayalon J., and Leichter I.Increased trabecular bone density due to bone-loading exercises in postmenopausal osteoporotic women. Calcif Tissue Int 40,59, 1987 [DOI] [PubMed] [Google Scholar]
- 25.Rui Y.F., Lui P.P.Y., Ni M., Chan L.S., Lee Y.W., and Chan K.M.Mechanical loading increased BMP-2 expression which promoted osteogenic differentiation of tendon-derived stem cells. J Orthop Res 29,390, 2010 [DOI] [PubMed] [Google Scholar]
- 26.Friedl G., Schmidt H., Rehak I., Kostner G., Schauenstein K., and Windhager R.Undifferentiated human mesenchymal stem cells (hMSCs) are highly sensitive to mechanical strain: transcriptionally controlled early osteo-chondrogenic response in vitro. Osteoarthritis Cartilage 15,1293, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Li Y.J., Batra N.N., You L., Meier S.C., Coe I.A., Yellowley C.E., and Jacobs C.R.Oscillatory fluid flow affects human marrow stromal cell proliferation and differentiation. J Orthop Res 22,1283, 2004 [DOI] [PubMed] [Google Scholar]
- 28.McCabe L.R., Kockx M., Lian J., Stein J., and Stein G.Selective expression of fos-and jun-related genes during osteoblast proliferation and differentiation. Exp Cell Res 218,255, 1995 [DOI] [PubMed] [Google Scholar]
- 29.Wagner E.Functions of AP1 (Fos/Jun) in bone development. Ann Rheumat Dis 61,ii40, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCabe L.R., Banerjee C., Kundu R., Harrison R.J., Dobner P.R., Stein J.L., Lian J.B., and Stein G.S.Developmental expression and activities of specific fos and jun proteins are functionally related to osteoblast maturation: role of Fra-2 and Jun D during differentiation. Endocrinology 137,4398, 1996 [DOI] [PubMed] [Google Scholar]
- 31.Yeo H., Beck L.H., McDonald J.M., and Zayzafoon M.Cyclosporin A elicits dose-dependent biphasic effects on osteoblast differentiation and bone formation. Bone 40,1502, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furlan F., Galbiati C., Jorgensen N.R., Jensen J.E.B., Mrak E., Rubinacci A., Talotta F., Verde P., and Blasi F.Urokinase plasminogen activator receptor affects bone homeostasis by regulating osteoblast and osteoclast function. J Bone Miner Res 22,1387, 2007 [DOI] [PubMed] [Google Scholar]
- 33.Häckel C., Radig K., Röse I., and Roessner A.The urokinase plasminogen activator (u-PA) and its inhibitor (PAI-1) in embryo-fetal bone formation in the human: an immunohistochemical study. Anat Embryol 192,363, 1995 [DOI] [PubMed] [Google Scholar]
- 34.Li X., Wang H., Touma E., Rousseau E., Quigg R.J., and Ryaby J.T.Genetic network and pathway analysis of differentially expressed proteins during critical cellular events in fracture repair. J Cell Biochem 100,527, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Ghosh-Choudhury N., Abboud S.L., Nishimura R., Celeste A., Mahimainathan L., and Choudhury G.G.Requirement of BMP-2-induced phosphatidylinositol 3-kinase and Akt serine/threonine kinase in osteoblast differentiation and Smad-dependent BMP-2 gene transcription. J Biol Chem 277,33361, 2002 [DOI] [PubMed] [Google Scholar]
- 36.Omasu F., Ezura Y., Kajita M., Ishida R., Kodaira M., Yoshida H., Suzuki T., Hosoi T., Inoue S., and Shiraki M.Association of genetic variation of the RIL gene, encoding a PDZ-LIM domain protein and localized in 5q31. 1, with low bone mineral density in adult Japanese women. J Hum Genet 48,342, 2003 [DOI] [PubMed] [Google Scholar]
- 37.Xiong Q., Jiao Y., Hasty K.A., Canale S.T., Stuart J.M., Beamer W.G., Deng H.W., Baylink D., and Gu W.Quantitative trait loci, genes, and polymorphisms that regulate bone mineral density in mouse. Genomics 93,401, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki M., Abe R., Fujita Y., Ando S., Inokuma D., and Shimizu H.Mesenchymal stem cells are recruited into wounded skin and contribute to wound repair by transdifferentiation into multiple skin cell type. J Immunol 180,2581, 2008 [DOI] [PubMed] [Google Scholar]
- 39.Li W.J., Tuli R., Okafor C., Derfoul A., Danielson K.G., Hall D.J., and Tuan R.S.A three-dimensional nanofibrous scaffold for cartilage tissue engineering using human mesenchymal stem cells. Biomaterials 26,599, 2005 [DOI] [PubMed] [Google Scholar]
- 40.Tuan R.S., Boland G., and Tuli R.Adult mesenchymal stem cells and cell-based tissue engineering. Arthritis Res Ther 5,32, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meinel L., Karageorgiou V., Fajardo R., Snyder B., Shinde-Patil V., Zichner L., Kaplan D., Langer R., and Vunjak-Novakovic G.Bone tissue engineering using human mesenchymal stem cells: effects of scaffold material and medium flow. Ann Biomed Eng 32,112, 2004 [DOI] [PubMed] [Google Scholar]
- 42.Yoshimoto H., Shin Y., Terai H., and Vacanti J.A biodegradable nanofiber scaffold by electrospinning and its potential for bone tissue engineering. Biomaterials 24,2077, 2003 [DOI] [PubMed] [Google Scholar]
- 43.Kadner A., Hoerstrup S.P., Zund G., Eid K., Maurus C., Melnitchouk S., Grunenfelder J., and Turina M.I.A new source for cardiovascular tissue engineering: human bone marrow stromal cells. Eur J Cardiothorac Surg 21,1055, 2002 [DOI] [PubMed] [Google Scholar]
- 44.Sumanasinghe R.D., Pfeiler T.W., Monteiro-Riviere N.A., and Loboa E.G.Expression of proinflammatory cytokines by human mesenchymal stem cells in response to cyclic tensile strain. J Cell Physiol 219,77–83, 2009 [DOI] [PubMed] [Google Scholar]
- 45.Tokalov S.V., Gruener S., Schindler S., Iagunov A.S., Baumann M., and Abolmaali N.D.A number of bone marrow mesenchymal stem cells but neither phenotype nor differentiation capacities changes with age of rats. Mol Cells 24,255, 2007 [PubMed] [Google Scholar]
- 46.Stolzing A., Jones E., McGonagle D., and Scutt A.Age-related changes in human bone marrow-derived mesenchymal stem cells: consequences for cell therapies. Mech Ageing Dev 129,163, 2008 [DOI] [PubMed] [Google Scholar]
- 47.Baxter M.A., Wynn R.F., Jowitt S.N., Wraith J.E., Fairbairn L.J., and Bellantuono I.Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem cells 22,675, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Muschler G.F., Nitto H., Boehm C.A., and Easley K.A.Age- and gender-related changes in the cellularity of human bone marrow and the prevalence of osteoblastic progenitors. J Orthop Res 19,117, 2001 [DOI] [PubMed] [Google Scholar]
- 49.Oreffo R., Bennett A., Carr A., and Triffitt J.Patients with primary osteoarthritis show no change with ageing in the number of osteogenic precursors. Scand J Rheumatol 27,415, 1998 [DOI] [PubMed] [Google Scholar]
- 50.Stenderup K., Justesen J., Clausen C., and Kassem M.Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone 33,919, 2003 [DOI] [PubMed] [Google Scholar]
- 51.Zhou S., Greenberger J.S., Epperly M.W., Goff J.P., Adler C., LeBoff M.S., and Glowacki J.Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging cell 7,335, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song G., Ju Y., Shen X., Luo Q., Shi Y., and Qin J.Mechanical stretch promotes proliferation of rat bone marrow mesenchymal stem cells. Colloids Surf B Biointerfaces 58,271, 2007 [DOI] [PubMed] [Google Scholar]
- 53.Kaspar D., Seidl W., Neidlinger-Wilke C., Beck A., Claes L., and Ignatius A.Proliferation of human-derived osteoblast-like cells depends on the cycle number and frequency of uniaxial strain. J Biomech 35,873, 2002 [DOI] [PubMed] [Google Scholar]
- 54.Li G., Simpson A., Kenwright J., and Triffitt J.Assessment of cell proliferation in regenerating bone during distraction osteogenesis at different distraction rates. J Orthop Res 15,765, 1997 [DOI] [PubMed] [Google Scholar]
- 55.Meyer U., Meyer T., Schlegel W., Scholz H., and Joos U.Tissue differentiation and cytokine synthesis during strain-related bone formation in distraction osteogenesis. Br J Oral Maxillofac Surg 39,22, 2001 [DOI] [PubMed] [Google Scholar]
- 56.Tang N., Zhao Z., Zhang L., Yu Q., Li J., Xu Z., and Li X.Up-regulated osteogenic transcription factors during early response of human periodontal ligament stem cells to cyclic tensile strain. Arch Med Sci 8,422, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung E., and Rylander M.N.Response of a preosteoblastic cell line to cyclic tensile stress conditioning and growth factors for bone tissue engineering. Tissue Eng Part A 18,397, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo Y., Zhang C., Zeng Q., Li R., Liu L., Hao Q., Shi C., Zhang X., and Yan Y.Mechanical strain promotes osteoblast ECM formation and improves its osteoinductive potential. Biomed Eng Online 11,1, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kasper G., Dankert N., Tuischer J., Hoeft M., Gaber T., Glaeser J.D., Zander D., Tschirschmann M., Thompson M., and Matziolis G.Mesenchymal stem cells regulate angiogenesis according to their mechanical environment. Stem Cells 25,903, 2007 [DOI] [PubMed] [Google Scholar]
- 60.Liu Y., Berendsen A.D., Jia S., Lotinun S., Baron R., Ferrara N., and Olsen B.R.Intracellular VEGF regulates the balance between osteoblast and adipocyte differentiation. J Clin Invest 122,3101, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Granchi D., Ochoa G., Leonardi E., Devescovi V., Baglìo S.R., Osaba L., Baldini N., and Ciapetti G.Gene expression patterns related to osteogenic differentiation of bone marrow–derived mesenchymal stem cells during ex vivo expansion. Tissue Eng Part C Methods 16,511, 2009 [DOI] [PubMed] [Google Scholar]
- 62.Xiao L., Yuan X., and Sharkis S.J.Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells. Stem Cells 24,1476, 2006 [DOI] [PubMed] [Google Scholar]
- 63.Westendorf J.J., Kahler R.A., and Schroeder T.M.Wnt signaling in osteoblasts and bone diseases. Gene 341,19, 2004 [DOI] [PubMed] [Google Scholar]
- 64.Lin G.L., and Hankenson K.D.Integration of BMP, Wnt, and Notch signaling pathways in osteoblast differentiation. J Cell Biochem 112,3491, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saraswati S., Bastakoty D., and Young P.P.Molecular and signaling pathways that modulate mesenchymal stem cell self-renewal. Stem Cells Cancer Stem Cells 6,131, 2012 [Google Scholar]
- 66.Paling N.R.D., Wheadon H., Bone H.K., and Welham M.J.Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. J Biol Chem 279,48063, 2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.