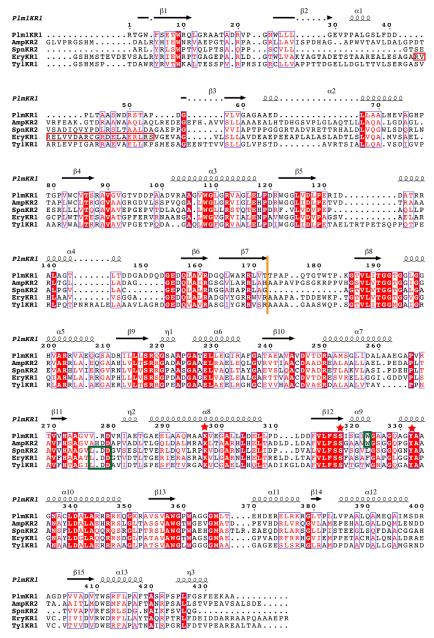
Figure 3. Structure based sequence alignment.
Structure-based multiple sequence alignment of PlmKR1 (accession AAQ84156), AmpKR2 (37% sequence identity to PlmKR1, accession AAK73513), SpnKR2 (32% sequence identity to PlmKR1, accession AAG23265), EryKR1 (35% identity to PlmKR1, accession AAB84070), TylKR1 (41% identity to PlmKR1, accession AAB66504). The red box indicates the location of β-strand and α-helix in EryKR1, which PlmKR1 lacks. Additionally, dotted lines in secondary structure elements indicate truncations of these elements in PlmKR1 when compared to the other KRs. Orange line indicates the end of the structural domain and start of the catalytic domain. Green boxes indicate the location of the W motif for A-type KRs (PlmKR1 and AmpKR2) and LDD motif for (B-type KRs). Red stars indicate the location of the active site residues. The sequence alignment was generated with MUSCLE (Edgar, 2004) and ESPript (Gouet et al., 1999) was used to generate secondary structure annotations.