This work identifies a Pooideae-specific microRNA that posttranscriptionally regulates the florigen gene FT under different daylength conditions in Brachypodium distachyon, revealing one mechanism in the complex but precise genetic regulatory pathways for flowering time control in plants.
Abstract
The highly conserved florigen gene FLOWERING LOCUS T (FT) functions at the core of the flowering pathways. Extensive studies have examined the transcriptional regulation of FT; however, other layers of FT regulation remain unclear. Here, we identified miR5200 a Pooideae-specific microRNA that is expressed in leaves and targets Brachypodium distachyon FT orthologs for mRNA cleavage. miR5200 was abundantly expressed in plants grown under short-day (SD) conditions but was dramatically repressed in plants transferred to long-day (LD) conditions. We also found that the epigenetic chromatin status, specifically the levels of histone methylation marks, at miR5200 precursor loci changed in response to daylength. Moreover, artificial interruption of miR5200 activity by target mimicry in B. distachyon altered flowering time in SD but not in LD conditions, suggesting that miR5200 functions in photoperiod-mediated flowering time regulation. Together, these findings illustrate a posttranscriptional regulation mechanism of FT and provide insights into understanding of the multiple concerted pathways for flowering time control in plants.
INTRODUCTION
The transition from vegetative growth to flowering stage is a critical event for seed propagation in plants. The correct timing of flowering initiation depends on five genetic pathways: photoperiod, vernalization, autonomy, hormones, and age (Fornara et al., 2010). These endogenous and environmental flowering regulatory cues ultimately regulate a set of integrator genes, such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), LEAFY, and FLOWERING LOCUS T (FT) (Kobayashi and Weigel, 2007).
Photoperiodic control of flowering in plants begins with the perception of daylength in leaves, followed by the transmission of a floral signal termed florigen into the shoot apex where the flowers are initiated (Bäurle and Dean, 2006). The mobile florigen signal is encoded by FT and FT orthologous genes in various plant species (Lifschitz et al., 2006; Tamaki et al., 2007; Pin et al., 2010; Meng et al., 2011; Navarro et al., 2011; Wigge, 2011). Mutation in FT causes a serious delay in flowering, whereas overexpression of FT induces precocious flowering, indicating that FT is necessary and sufficient to accelerate the floral transition (Kardailsky et al., 1999; Kobayashi et al., 1999). In Arabidopsis thaliana, FT protein travels from the vasculature to the shoot apex to form an effector complex with FD, a bZIP transcription factor, and thereby stimulates flower organ formation through activation of a subset of downstream floral genes, including APETALA1 (AP1), FRUITFULL (FUL), and SOC1 (Abe et al., 2005; Wigge et al., 2005; Corbesier et al., 2007; Mathieu et al., 2007).
Positive and negative regulators control FT transcription. Under long-day (LD) conditions, FT expression is mainly activated in vascular bundles via transcriptional regulators CONSTANS (CO) and GIGANTEA (Suárez-López et al., 2001; Sawa and Kay, 2011; Torti et al., 2012). CO is directly recruited to the FT promoter or indirectly acts with other transcriptional cofactors to regulate FT expression (Wenkel et al., 2006; Kobayashi and Weigel, 2007). The age-dependent SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 can also directly act at the FT promoter and modulate ambient temperature-responsive flowering (Wang et al., 2009; Wu et al., 2009a; Kim et al., 2012). As a major negative regulator of FT, FLOWERING LOCUS C forms a MADS box complex with SHORT VEGETATIVE PHASE, which directly inhibits FT expression to mediate the effects of exposure to cold conditions or ambient temperature in Arabidopsis (Bastow et al., 2004; Bäurle and Dean, 2006; Li et al., 2008). Furthermore, two TEMPRANILLO proteins (TEM1 and TEM2) directly repress FT and may have antagonistic roles with the activator CO to determine FT levels (Castillejo and Pelaz, 2008). Several miR172-targeted AP2-like genes also affect the repression of FT transcription, but their modes of action remain unclear (Mathieu et al., 2009).
In addition to transcription factors, trimethylation of H3K27 (H3K27me3) and demethylation of H3K4 (H3K4me2) modifications in cis also epigenetically regulate FT transcription (Farrona et al., 2008; Adrian et al., 2010). FT might also be controlled at posttranscriptional levels, since the werewolf root hair patterning mutant indirectly affected FT mRNA stability in Arabidopsis, but the molecular basis of this process is still elusive (Seo et al., 2011).
Compared with eudicots, the behaviors of FT during floral induction in monocots are largely unknown to date. In rice (Oryza sativa), a typical short-day (SD) plant, two FT-like proteins referred to as Heading-date3a and RICE FLOWERING LOCUS T1, which are respectively activated under SD and LD conditions, have been proposed to be the SD- and LD-specific florigens (Komiya et al., 2009). Ectopic expression of the FT-like gene VERNALIZATION3 (VRN3) in wheat (Triticum aestivum) and barley (Hordeum vulgare) activated transcription of the AP1 orthologous gene VRN1 and promoted early flowering (Trevaskis et al., 2003; Yan et al., 2006).
With its rapid growth, small genome, and simple growth conditions, Brachypodium distachyon has been thought as a suitable model system for studies on temperate cereals and biofuel plants, such as switchgrass (Panicum virgatum; Opanowicz et al., 2008); however, so far, information about its flowering is quite limited (Higgins et al., 2010; Hong et al., 2010). B. distachyon accessions show substantial natural variation in flowering time. For example, the genome-sequenced accession Bd21 is a spring annual and can flower without any vernalization, whereas the winter accession Bd1-1 requires vernalization, a period of cold treatment, for flowering (Schwartz et al., 2010). In addition, daylength greatly influences the flowering time of B. distachyon accessions. For instance, Bd21 can flower within 45 d of growth under LD conditions but will delay flowering for half a year if grown in SD environments (Schwartz et al., 2010; see Supplemental Figures 1A and 1B online). Thus, there may be special components that precisely regulate flowering time in B. distachyon.
The 21- to 24-nucleotide microRNAs (miRNAs), a class of DICER-LIKE (DCL)–dependent small RNAs (sRNAs), regulate a wide range of biological processes in monocots and eudicots (Voinnet, 2009; Wu et al., 2009b). In this study, we characterized a Pooideae-specific miR5200 that guides sequence-specific cleavage of FT orthologous transcripts in B. distachyon leaves. We further demonstrate that miR5200 plays an important role in regulation of the photoperiod-mediated floral transition. Our findings show that a miRNA directly mediates FT posttranscriptional modulation in Pooideae plants, which may also supply an alternative approach for heading date breeding in barley and wheat.
RESULTS
Hairpin Structures and Primary Transcripts of miR5200
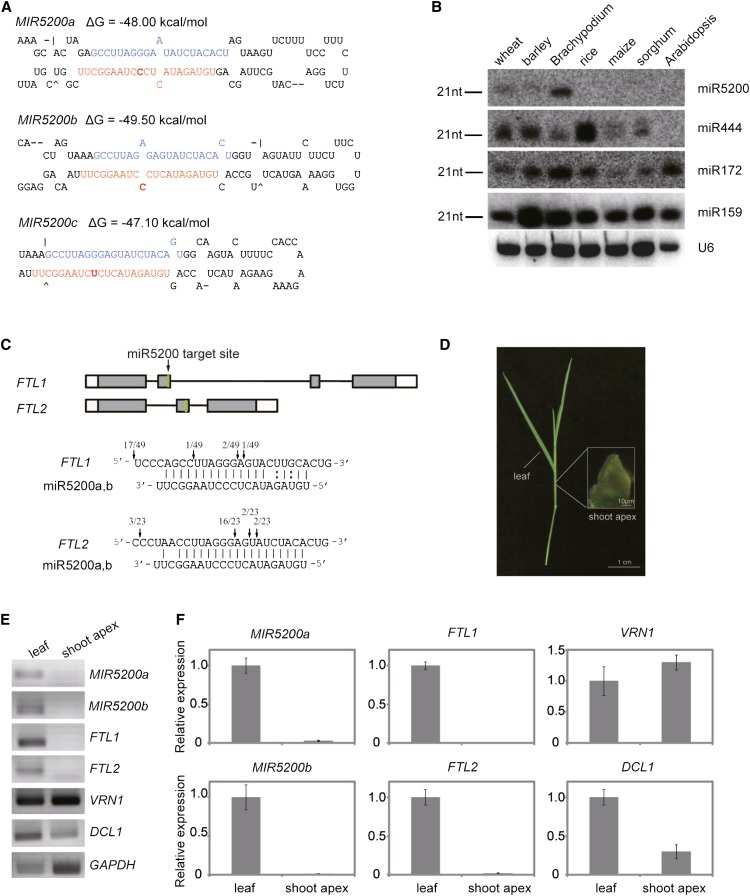
We previously identified a subset of miRNAs by deep sequencing of small RNAs in bread wheat. One 21-nucleotide miRNA, which we named miR2032, was identified, and its targets were predicted in silico to be FT orthologous genes (Wei et al., 2009). We found that another miRNA, previously annotated in miRBase as miR5200 in B. distachyon (Zhang et al., 2009), has only one nucleotide difference with the miRNA we named miR2032 in bread wheat (Wei et al., 2009). In addition, miR2032 has been assigned to another miRNA in sea anemone in miRBase. Therefore, we have renamed this miRNA miR5200a, and the sequence identified by Zhang et al. (2009) as miR5200c. Owing to the importance of FT-like proteins in plant flowering pathways, we further analyzed this miRNA in depth. To determine the conservation of miR5200 in other plant species, we performed a BLAST search of all available small RNA databases. We only detected the 21-nucleotide miR5200 sequence in the sRNA data sets of a number of Pooideae species, including barley and B. distachyon (Schreiber et al., 2011; Hackenberg et al., 2012).
Since B. distachyon has emerged as a research model for Pooideae plants, by virtue of its published high-quality genome sequence and tractable genetic transformation systems (Vogel et al., 2010; Brkljacic et al., 2011), we investigated the structure and functions of miR5200 in detail in B. distachyon.
Bioinformatic prediction of miR5200a stem-loop structures found that two loci on B. distachyon chromosome 1 potentially encoded the miR5200a transcribed genes, and we named these two genes MIR5200a and MIR5200b (Figure 1A). Interestingly, like in wheat, B. distachyon MIR5200c and MIR5200b were located at the same but reverse complementary loci, suggesting that both mature sequences and precursor structures of miR5200 are conserved between B. distachyon and wheat (Abrouk et al., 2012; Lucas and Budak, 2012). RNA gel blot analysis validated that miR5200 was present in the three Pooideae plants, but not in rice (Oryza sativa), maize (Zea mays) or sorghum (Sorghum bicolor), nor in dicotyledonous plants such as Arabidopsis thaliana (Figure 1B). These results suggest that miR5200 is a recently evolved and Pooideae-specific miRNA (see Supplemental Figures 2A and 2B).
Figure 1.
Hairpin Structures, Expression Patterns, and Target Validation of miR5200.
(A) The 5′ to 3′ secondary structures of pre-miR5200 predicted for B. distachyon. The mature miR5200 and miR5200* sequences are indicated in red and blue, respectively. The different nucleotides between miR5200a and b/c are highlighted by bold letters.
(B) RNA gel blot analysis of miR5200 in wheat, barley, B. distachyon, rice, maize, sorghum, and Arabidopsis. Note that miR444 is a monocot-specific miRNA, whereas miR172 and miR159 are conserved between monocots and dicots. U6 was used as RNA loading control. nt, nucleotides.
(C) Schematic representation of gene structure and validation of miR5200 targets. Gene exons, introns, and target regions are shown by rectangles, lines, and green bars, respectively. Arrows indicate the cleavage sites detected by 5′-RACE.
(D) Parts of leaf and shoot apex in B. distachyon that were dissected for miR5200 spatial expression analysis.
(E) Spatial expressions of miR5200 and target genes in leaf and shoot apex. The expression of pri-miR5200 and indicated genes was analyzed by RT-PCR with amplification of GAPDH mRNA as a control.
(F) qPCR analysis of pri-miR5200 and indicated gene expression in leaf and shoot apex. GAPDH was used as an internal control for normalization of qPCR results. Each point represents the average of three biological replicates, and error bars indicate sd.
To better understand the complete gene structures of primary miR5200 (pri-miR5200), we used RNA ligase-mediated rapid amplification of cDNA ends (RACE) to retrieve 5′ and 3′ sequences of the transcript. Like canonical miRNAs (Xie et al., 2005), we found that both MIR5200a and MIR5200b transcripts were 5′ capped with 3′ polyadenylation (see Supplemental Figures 3A and 3B online). Intriguingly, we detected 14 splice variants of MIR5200a, but each variant had the same 5′ segment (see Supplemental Figure 3A online). Distinct from various MIR5200a transcripts that carried one to several introns, the short MIR5200b transcript had only one exon (see Supplemental Figure 3B online).
miR5200 Targets FT Orthologous Genes and Accumulates in Leaves
Similar to the initial computational prediction of miR5200a targets in wheat and barley (see Supplemental Figures 4A and 4B online), using psROBOT sRNA analysis toolbox (Wu et al., 2012), we identified two B. distachyon FT-like genes (Bradi2g07070 and Bradi1g48830), which were previously annotated as FTL1 and FTL2 (Higgins et al., 2010), as miR5200a targets (Figure 1C; see Supplemental Figures 4C and 4D online). Further validation through 5′ RNA ligase-mediated RACE showed that both FTL1 and FTL2 mRNAs could be cleaved at miR5200 target sites in vivo, suggesting that these two FTs are bona fide miR5200 target genes (Figure 1C). Although a majority of the cleavage events of FTL1 occurred outside of the canonical miRNA cleavage site, it is unlikely that this cleavage was mediated by secondary siRNAs because miR5200a is not 22 nucleotides in length and there are no bulged bases between the miRNA:miRNA* (for miRNA star) duplex that are known to be required for secondary siRNA formation (Chen et al., 2010; Cuperus et al., 2010; Manavella et al., 2012).
Since FT is not expressed at the shoot apex, it is reasonable to ask whether miR5200 is also specifically expressed outside the shoot apex to degrade FT mRNAs. To address this question, we collected B. distachyon leaves and shoot apices and performed RT-PCR to determine MIR5200 expression patterns in these tissues (Figure 1D). Strikingly, we found that both MIR5200a and MIR5200b dramatically accumulated in leaves, but neither of them appeared in shoot apex (Figures 1E and 1F). By contrast, AP1 ortholog VRN1 and GAPDH genes were present in both tissues (Figures 1E and 1F). Together with the observation that DCL1 was expressed in both leaves and shoot apices, these results suggest that miR5200 is specifically expressed in leaves to repress FT mRNAs.
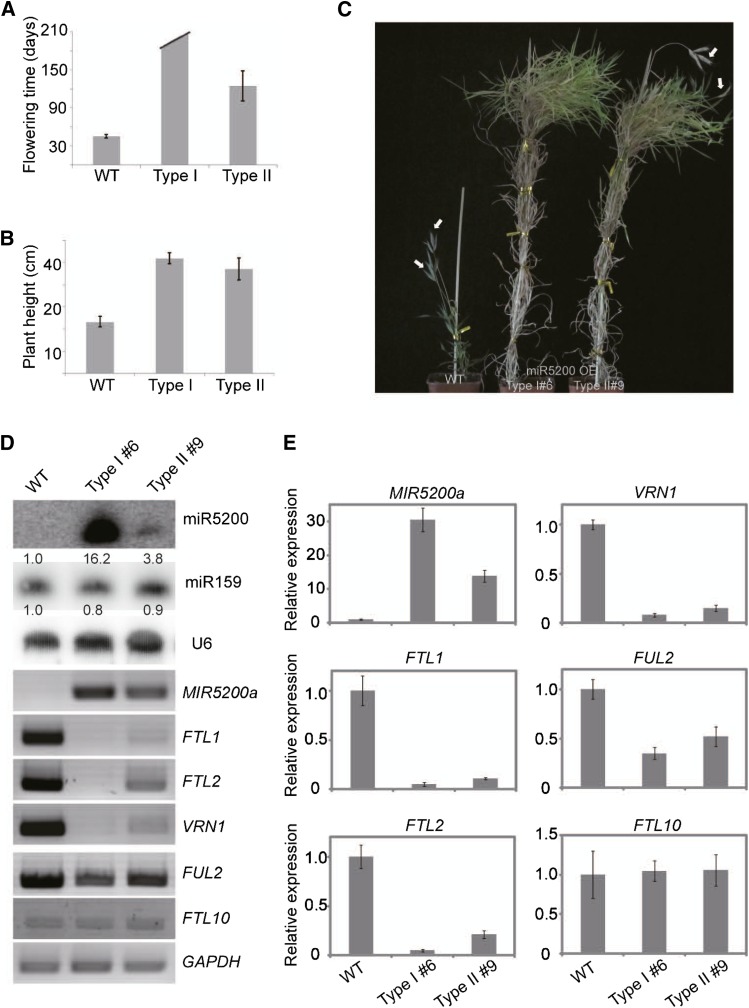
To verify that miR5200 targets FTs in vivo, we generated miR5200 overexpression (miR5200-OE) transgenic B. distachyon plants to trace whether FTLs were inhibited when miR5200 increased. We obtained 11 independent transgenic lines from separately generated calluses and all of them showed delayed flowering time phenotypes in LD environments (see Supplemental Figures 5A and 5B online). Based upon the severity of late flowering time, these lines were divided into Types I and II (Figure 2A): Type I did not flower even after 180 d and thus could not form seeds; Type II bolted much later than the wild type (110 versus 45 d) and produced fewer seeds than the wild-type plants (Figures 2A and 2C). Moreover, both types of miR5200-OE plants eventually grew much higher than the wild type (Figures 2B and 2C). RT-PCR showed that the expression of FTL1 and FTL2 was dramatically reduced in Types I and II transgenic lines, whereas that of another FT-like gene (FTL10) without the miR5200 target site was comparable to the wild type (Figures 2D and 2E). We also found that B. distachyon AP1 and FUL orthologous genes, VRN1 and FUL2, which may act downstream of FT (Li and Dubcovsky, 2008), also displayed decreased mRNA levels in miR5200-OE plants (Figures 2D and 2E). These results indicate that miR5200 mediates FTL1 and FTL2 mRNA degradation for posttranscriptional regulation.
Figure 2.
miR5200 Overexpression Severely Delayed Flowering Time in Transgenic B. distachyon Plants.
(A) Flowering time of wild-type (WT) Bd21 and the indicated two types of miR5200-OE transgenic T0 plants. Three plants for type I and eight plants for type II were scored. Error bars indicate sd.
(B) Plant heights of the wild type and two types of miR5200-OE transgenic T0 plants. Three type I plants and eight type II plants were measured. Error bars indicate sd.
(C) Phenotypes of ectopic miR5200 expression T0 transgenic B. distachyon plants in LDs. The left plant is 2-month-old wild type. The middle and right plants are representatives of 4-month-old miR5200-OE transgenic plants with strong and weak phenotypes respectively. White arrows point to spikes.
(D) miR5200 (by RNA gel-blots) and flowering-time genes (by RT-PCR) expression in wild-type and the indicated miR5200-OE transgenic plants. Numbers below each miRNA gel blot denote fold changes relative to the miRNA level in wild-type plants.
(E) qPCR analysis of pri-miR5200 and indicated flowering gene expressions in wild-type and the indicated miR5200-OE transgenic plants. GAPDH was used as an internal control for normalization of qPCR results. Each point represents the average of three technical replicates, and error bars indicate sd.
[See online article for color version of this figure.]
Photoperiod Affects miR5200 Expression
The initiation of flowering in temperate grasses depends on delicate regulation of FT, which plays a central role in balancing competing signals from vernalization and photoperiod (Distelfeld et al., 2009). To determine whether miR5200 was integrated in FT regulation in B. distachyon flowering, we examined the expression patterns of miR5200 in response to vernalization and daylength.
Treating B. distachyon accession Bd21 under 4°C low temperature for 2 to 4 weeks can accelerate plant flowering, but we observed that mature miR5200 did not change in 2- or 4-week cold-treated Bd21 plants (see Supplemental Figure 6A online), suggesting that vernalization may not affect miR5200 in Bd21. Bd1-1 is a Turkish winter-habit B. distachyon accession and cannot flower without vernalization (Filiz et al., 2009). However, 2 to 4 weeks of cold treatment did not alter the abundance of miR5200 in Bd1-1 either (see Supplemental Figure 6A online). These results reveal that the regulation of FT by miR5200 is likely to be independent of the vernalization pathway in B. distachyon.
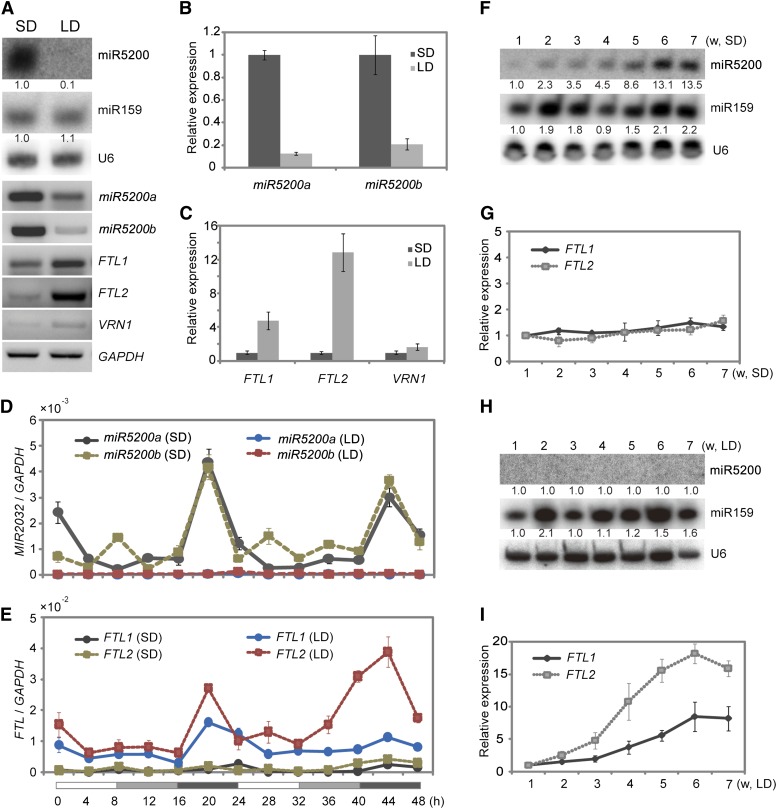
Next, we examined the amounts of miR5200 in plants grown under different daylengths. As shown in Figure 3A, RNA gel blots showed that miR5200 accumulated to high levels in 8-h-light SD conditions but was nearly undetectable in 16-h-light LD conditions. RT-PCR and quantitative real-time PCR (qPCR) analyses also revealed that the primary transcripts of MIR5200a and MIR5200b were much higher in SDs than in LDs (Figures 3A to 3C). These results suggest that miR5200 may be controlled by the photoperiod. To further assess the reliance of miR5200 on daylength, we detected the abundance of miR5200 under 4-, 8-, 16-, and 20-h-light conditions. We found that miR5200 had similar levels under 4- and 8-h-light SD conditions, much higher than in 16- and 20-h-light LDs (see Supplemental Figure 6B online).
Figure 3.
Photoperiod Affects miR5200 Expression.
(A) Mature miR5200 (by RNA gel blots), pri-miR5200, and flowering-time genes (by RT-PCR) expression under SDs and LDs. The numbers below each miRNA gel blot denote fold changes relative to the miRNA level under SDs.
(B) qPCR analysis of pri-miR5200 in B. distachyon plants under SDs and LDs.
(C) qPCR analysis of the indicated flowering gene expressions in SDs and LDs.
(D) Diurnal expression patterns of MIR5200a and MIR5200b in B. distachyon under SDs and LDs.
(E) Diurnal expression patterns of FTL1 and FTL2 under SDs and LDs. Each point represents the average of three technical replicates, and the error bars indicate repeat sd. The white and black bars along the horizontal axes represent light and dark periods, respectively. The numbers below the horizontal axes indicate the time in hours.
(F) and (G) Growth stage–dependent expressions of miR5200 (F) and its target genes (G) in SDs. Numbers below each miRNA gel blot indicate fold changes relative to the miRNA level in 1-week SD plants. w, weeks.
(H) and (I) Growth stage–dependent abundance of miR5200 (H) and its target gene expression (I) under LDs. Each point represents the average of three biological replicates, and error bar indicates sd. GAPDH was used as an internal control for normalization of qPCR results, and U6 was served as loading control for RNA gel blots. Numbers below each miRNA gel blot indicate the fold changes relative to the miRNA level in 1-week-old plants under LDs.
Considering that FT family genes usually showed diurnal expression rhythms, we examined the oscillation of pri-miR5200 and targets every 4 h during the day-night cycle in B. distachyon. Under SDs, both MIR5200a and MIR5200b displayed an expression peak at 4 h before dawn (Figure 3D), and FTLs also peaked at this time but in LDs (Figure 3E). Furthermore, as shown in Figures 3D and 3E, MIR5200a and MIR5200b were expressed at much higher levels, whereas FTL1 and FTL2 accumulated to much lower levels at any time point in SDs than LDs during the 48-h time-course examination.
In Arabidopsis, FT mRNA levels gradually increased with growth time, especially under LD conditions (Kobayashi et al., 1999). Therefore, we asked whether the abundance of miR5200 changed progressively with FT mRNA increases under SDs and LDs in B. distachyon. Under SDs, FTL1 and FTL2 expression levels remained constant, whereas miR5200 showed a clear increase along with growth time (Figures 3F and 3G). Under LDs, both FTL1 and FTL2 showed a gradual increase in expression along with the photoperiod time; however, we did not detect miR5200 expression even after 7 weeks (Figures 3H and 3I). These results suggest that miR5200 appears to specifically inhibit FTL1 and FTL2 accumulation in SDs.
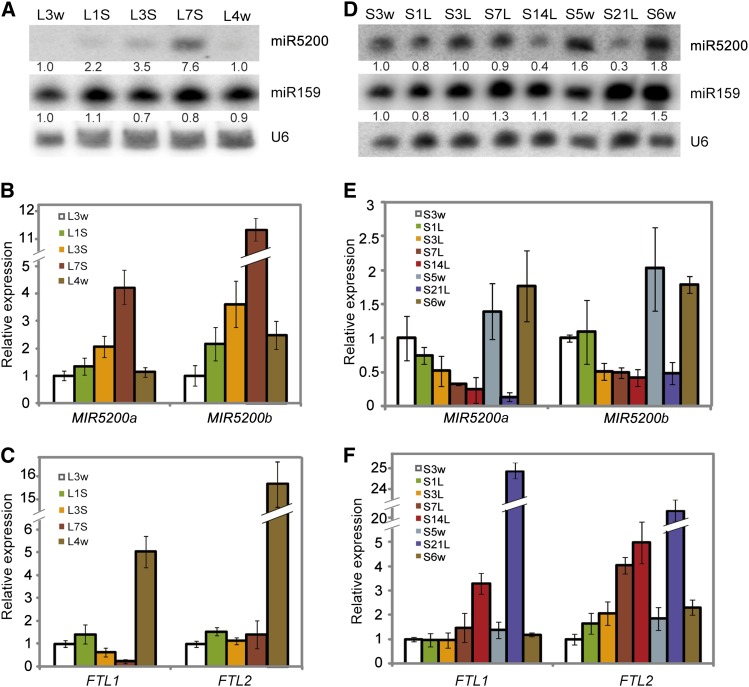
High expression in SDs and low abundance of miR5200 in LDs as described above implied that photoperiod may affect FT expression through regulation of miR5200. To further pursue the effects of daylength on miR5200 and its targets, we examined the dynamic changes of miR5200 by RNA gel blots, shifting plants from LDs to SDs and vice versa. When LD-grown plants were moved from LDs to SDs for 1 d, both pri-miR5200 and mature miR5200 were elevated and gradually accumulated to a high level in 7 d (Figures 4A and 4B). In the meantime, FTL1 progressively decreased to a low level during this period (Figure 4C), suggesting a negative correlation between FTL1 and miR5200 in this process. When these plants were returned to SDs for another 7 d, FTL1 and FTL2 displayed a moderate increase along with a reduction of miR5200 (see Supplemental Figures 6C and 6D online). By contrast, the abundance of mature miR5200 in SD-grown plants remained constant after transfer to LDs for 7 d even as the pri-miRNA expression began to decline within 3 d (Figures 4D and 4E). When this shift time was extended to over 14 d, a gradual reduction of miR5200 was detected (Figure 4D). Interestingly, when these shifted plants were transferred back to SD conditions for another 7 d, miR5200 expression was recovered (see Supplemental Figure 6E online). Meanwhile, FTL1 and FTL2 displayed roughly complementary alterations during these shifts (Figure 4F; see Supplemental Figure 6F online), suggesting that these two FTLs are photoperiodically regulated by both miR5200 and additional flowering factors in response to daylength variation.
Figure 4.
Dynamic Effects of Daylength Changes on miR5200, FTL1, and FTL2 Accumulation.
(A) Time course of miR5200 expression when B. distachyon plants were shifted from LDs to SDs. L3w and L4w indicate that B. distachyon were grown under LDs for 3 and 4 weeks. L1S, L3S, and L7S represent 3-week-old plants that were moved from LDs to SDs for 1, 3, and 7 d, respectively. Numbers below each miRNA gel blot indicate fold changes relative to the miRNA level in wild-type Bd21 plants grown under LDs for 3 weeks.
(B) qPCR examination of dynamic changes of MIR5200a and MIR5200b in (A).
(C) qPCR analysis of dynamic effects of daylength on FTL1 and FTL2 in (A).
(D) Time course of miR5200 expression when 3-week-old SD plants were transferred to LDs for 1, 3, 7, 14, and 21 d. Plants grown under SDs for 5 and 6 weeks were harvested and used as controls. Numbers below each miRNA gel blot denote fold changes relative to the miRNA level in Bd21 grown under SDs for 3 weeks.
(E) qPCR examination of dynamic changes of MIR5200a and MIR5200b in (D).
(F) qPCR analysis of dynamic effects of daylength on FTL1 and FTL2 in (D). Error bar means sd (n = 3). Samples for RNA extraction in the experiment were collected at Zeitgeber time 2. U6 was used as loading control for RNA gel blots. GAPDH was used as an internal control for normalization of qPCR data.
Changes of Chromatin Structures at Pri-miR5200 Loci Result in Photoperiodic Expression of miR5200
Epigenetic mechanisms regulate numerous genes in flowering pathways, including Polycomb repressive complex inhibition of FT transcription (Farrona et al., 2008). To explore whether the transcriptional regulation of miR5200 by photoperiod was the result of epigenetic control, we first examined the status of DNA methylation at pri-miR5200 loci under SD and LD conditions by Chop-PCR (see Supplemental Figures 7A and 7B online). We observed no obvious differences of DNA methylation at MIR5200a and MIR5200b when B. distachyon was grown in SDs and LDs (see Supplemental Figures 7C and 7E online). Bisulfite sequencing of three regions confirmed these results (see Supplemental Figures 7D and 7F online), suggesting that the distinct patterns of MIR5200 transcription under different light conditions are not triggered by DNA methylation.
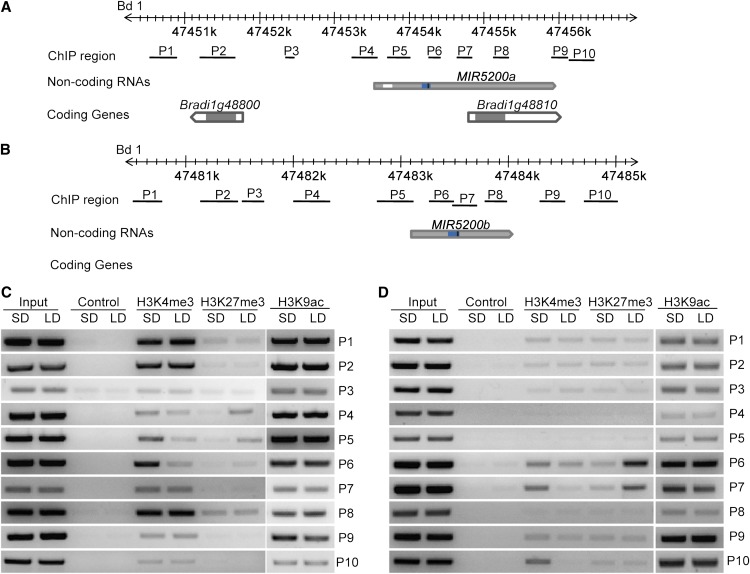
We then tested histone epigenetic status at MIR5200 loci under SDs and LDs using chromatin immunoprecipitation (ChIP) assay. We designed a series of oligos at both MIR5200 genes and their neighboring regions (Figures 5A and 5B). Although the enrichment of positive histone mark H3K9ac around MIR5200a and MIR5200b loci was similar under SDs and LDs (Figures 5C and 5D), we found that the repressive histone mark H3K27me3 surrounding the predicted hairpin structure regions of MIR5200 genes was enriched much higher under LDs than that under SDs (Figures 5C and 5D), suggesting that the suppression of pri-miR5200 transcripts under LDs is caused by the high level of H3K27me3 enrichment. H3K4me3 has been reported to act as bivalent mark together with H3K27me3 at the FT loci in Arabidopsis (Adrian et al., 2010). Our ChIP data showed that compared with those grown in LDs, H3K4me3 around the MIR5200a and MIR5200b stem-loop area was much enhanced when B. distachyon plants were grown under SDs (Figures 5C and 5D). Furthermore, we found that H3K4me3 was also highly enriched in the region of MIR5200b 1-kb terminator under SDs, despite that similar H3K27me3 status was found at this site in both SDs and LDs (Figure 5D).
Figure 5.
Histone Modification Status at Pri-miR5200 Loci in B. distachyon Grown under Different Daylengths.
(A) and (B) Diagrams showing the genomic locations, the adjacent coding genes, and the regions examined by ChIP assay for MIR5200a (A) and MIR5200b (B). Gray boxes represent exons, and white boxes represent introns. Blue boxes located in gray boxes indicate predicted MIR5200a and MIR5200b hairpin structures. Positions of mature miR5200 are set as black bars. ChIP regions are shown by black lines.
(C) and (D) Relative abundance of H3K4me3, H3K27me3, and H3K9ac at MIR5200a (C) and MIR5200b (D) genomic regions in B. distachyon grown under SDs and LDs. Note that the high levels of H3K4me3 and the decreased H3K27me3 around regions corresponding to MIR5200a (P4/P5/P6) and MIR5200b (P6/P7) hairpin structures are associated with the activation of MIR5200a and MIR5200b expression under SD conditions.
Taken together, our results indicate that MIR5200a and MIR5200b genomic regions are subject to changes in histone modification under different daylengths. These results suggest that the distinct expression of miR5200 for photoperiodic regulation of flowering under SDs and LDs is probably caused by epigenetic chromatin alterations at MIR5200 loci.
Disruption of miR5200 Activity Specifically Alters Flowering Time in SDs
miR5200 functions in photoperiodic modulation of FTs; therefore, one interesting question is what the consequences are if the miR5200 regulatory activity is compromised. To address this issue, we first attempted to generate transgenic lines that ectopically expressed FTL1 and FTL2 as well as their miRNA-resistant mutant forms FTL1m and FTL2m, which were resistant to interaction with miR5200 (see Supplemental Figures 8A, 8B, and 8D online). Unfortunately, we were unable to generate such transgenic plants, as all of them flowered extremely early before they could form roots in the regeneration medium (see Supplemental Figures 8C and 8E online). These results indicate that both FTL1 and FTL2 have florigen activity in B. distachyon.
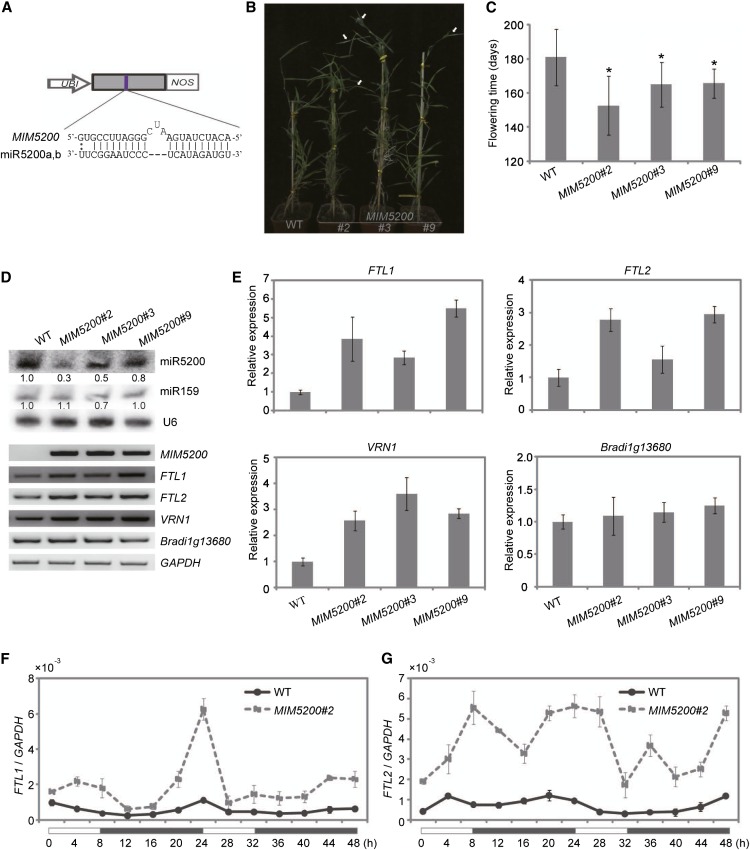
Artificial miRNA target mimics, RNA fragments that have similar sequence as the miRNA target but cannot be cleaved, can deplete miRNAs and thus reduce their function, providing useful reverse genetic tools to investigate biological functions of individual miRNA families in diverse plants (Franco-Zorrilla et al., 2007; Todesco et al., 2010; Bergonzi et al., 2013; Zhou et al., 2013). To gain more insights into photoperiodic regulation of flowering by miR5200, we designed and generated artificial target mimics for the miR5200 family and then expressed them in B. distachyon (Figure 6A). We termed these target mimic transgenic plants as MIM5200.
Figure 6.
Disruption of miR5200 Activity in B. distachyon Promotes Flowering under SDs.
(A) A diagram of MIM5200 showing the designed sequences for miR5200 target mimics. UBI, maize ubiquitin promoter; NOS, nos terminator.
(B) Representative photograph of flowering phenotypes in 6-month-old wild-type (WT) and indicated T1 MIM5200 transgenic plants grown in SDs. The white arrows point to spikes.
(C) Flowering time of wild-type and the indicated T1 MIM5200 transgenic lines grown in SDs. Error bar shows sd (n = 8). Asterisks indicate a significant difference between wild-type and transgenic plants (Student’s t test, P value < 0.05).
(D) Expression of miR5200 (by RNA gel blots), targets, and other flowering genes (by RT-PCR) as well as Bradi1g13680 (miR169 target gene as a control) in wild-type and the indicated T1 MIM5200 transgenic lines under SDs. U6 and GAPDH were served as the RNA gel blot and RT-PCR RNA loading controls, respectively. The numbers below each miRNA gel blot denote the fold changes relative to the miRNA level in the wild type.
(E) qPCR analysis of miR5200 target genes, VRN1, and Bradi1g13680 (miR169 target as a control) expression in wild-type and the indicated MIM5200 transgenic plants under SDs.
(F) and (G) Diurnal expression patterns of FTL1 (F) and FTL2 (G) in wild-type and the indicated MIM5200 transgenic plants under SDs. Each point represents the average of three technique replicates, and the error bar indicates sd. White and black bars along the horizontal axes represent light and dark periods, respectively. Numbers below the horizontal axes indicate the time in hours.
As miR5200 was expressed much higher under SDs than LDs, we first examined the flowering dates of MIM5200 plants in SDs. We observed that six out of nine independent MIM5200 T1 positive lines flowered 10 to 30 d earlier than wild-type plants when they were grown in SDs (Figures 6B and 6C), whereas the T1 segregated negative plants did not show any early heading phenotypes compared with the wild-type plants (see Supplemental Figures 9A and 9B online). To illustrate that the phenotypes of MIM5200 plants in SDs were indeed caused by the attenuated miR5200 activities, three independent lines were subject to further molecular analysis. As shown in Figures 6D and 6E, the expression of miR5200 was reduced in MIM5200 plants, while FTL1 and FTL2 as well as their downstream gene VRN1 increased. Moreover, we detected the diurnal expression patterns of FTL1 and FTL2 at 4-h intervals under SDs and found they exhibited significantly higher expression in MIM5200 than wild-type plants (Figures 6F and 6G). These findings strongly suggest that these two FT genes in B. distachyon are indeed regulated by miR5200 to make plants flower at the correct time in SDs.
To determine whether disruption of miR5200 activities in B. distachyon can affect FTs under LD conditions, we extracted total RNA from LD-grown wild-type and MIM5200 plants and measured the transcripts of FTLs. We did not detect obvious differences in expressions of FTL1 and FTL2 or VRN1, indicating that interruption of miR5200 activities in LDs may not affect FTs (see Supplemental Figures 9C and 9D online). Consistent with these results, the diurnal expression of FTL1 and FTL2 was similar in MIM5200 and wild-type plants under LDs (see Supplemental Figures 9E and 9F online). Furthermore, we observed that heading dates were nearly the same for wild-type and MIM5200 plants under LD environments (see Supplemental Figures 9G and 9H online). These results imply that miR5200 has little effect on regulation of B. distachyon flowering time in LD conditions.
Taken together, specific alteration of bolting time of MIM5200 plants in SDs suggests that miR5200 plays an important role in photoperiod-mediated flowering time regulation.
Photoperiodic Regulation of miR5200 Appears to Be Prevalent in the Pooideae
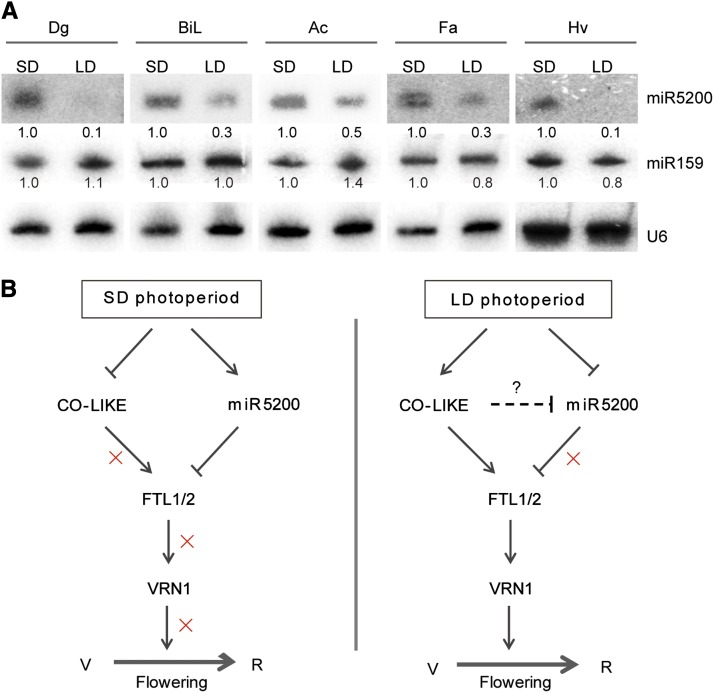
Considering the roles of miR5200 in B. distachyon flowering control stated above, we asked whether the photoperiodic modulation of miR5200 was conserved among species in the Pooideae. To address this question, we selected five Pooideae: Dactylis glomerata, Bromus inermis Leyss, Agropyron cristatum, Festuca arundinacea, and barley to compare miR5200 expression under SD and LD environments. As shown in Figure 7A, the abundance of miR5200 in all five grasses was much higher when they were grown under SDs than LDs, suggesting that photoperiodic regulation of miR5200 may have conserved roles in flowering time control among Pooideae plants.
Figure 7.
Photoperiodic Regulation of miR5200 Is Prevalent in Pooideae Plants.
(A) RNA gel blot analyses of miR5200 expression in selected Pooideae grasses under SDs and LDs. Dg, D. glomerata; BiL, B. inermis Leyss; Ac, A. cristatum; Fa, F. arundinacea; Hv, H. vulgare. U6 served as the RNA loading control of the RNA gel blot. Numbers below each miRNA gel blot represent the fold changes relative to the miRNA under SDs.
(B) A working model for miR5200 in the photoperiodic flowering pathway in Pooideae plants. B. distachyon is used here as an example. When B. distachyon grows under SDs, the repression of positive photoperiodic regulators (e.g., CO-like protein) as well as the activation of miR5200 lead to low transcripts of florigen genes, FTL1/2, and the downstream gene VRN1 and thereby cause late flowering. When B. distachyon grows under LDs, miR5200 is robustly inhibited, while the positive photoperiodic regulators (e.g., CO-like protein) are induced, resulting in high expression of FTL1/2, VRN1, and early flowering. The question mark indicates that it remains to be further elucidated whether positive regulators (e.g., CO-like protein) in the photoperiod pathway directly repress miR5200 activity under LDs. Lines with an arrow indicate positive regulation; lines with a vertical bar at the end represent repressive regulation. Crosses (x) mean the breakdown of the pathway. V, vegetative stage; R, reproductive stage.
[See online article for color version of this figure.]
DISCUSSION
Plants have evolved sophisticated ways to restrict flowering to the right time. As the most important flowering integrator, the florigen FT has been the focus of attention for many plant scientists. Their findings about FT regulation so far have centered on transcriptional regulation of FT; other layers of FT modulation, such as mRNA and protein stability, remain a puzzle. Here, we systemically characterized a newly evolved miR5200 in Pooideae subfamily plants and found that miR5200 functions in photoperiodic regulation of flowering time via cleavage of transcripts of FT-like genes.
MiRNAs function as key players in various aspects of plant biological processes (Voinnet, 2009); however, research on plant miRNA functions has mostly focused on conserved miRNAs, and the biological behaviors of nonconserved miRNAs, especially in monocots, had remained largely unknown (Chuck et al., 2009). The actions of miR5200 illustrated here may fill this vacancy to some extent. Even if FT genes have close relationships in rice and B. distachyon, there is no miR5200 counterpart in rice (Figure 1). This would not be a surprise, since Early heading date1, a central gene essential for photoperiodic regulation of flowering in rice, does not have an ortholog in B. distachyon either (Doi et al., 2004; Higgins et al., 2010). These phenomena suggest that each plant species may have acquired its own specific coding or noncoding RNAs to fine-tune the timing of flowering.
Several lines of evidence assessed here indicate that there are two florigen genes acting redundantly in B. distachyon: first, FTL1 and FTL2 overexpressing transgenic plants flowered early and arrested in the regeneration medium (see Supplemental Figure 8 online); second, FTL1 and FTL2 show over 79% amino acid sequence identity (see Supplemental Figure 4 and Supplemental Data Set 1 online); third, both FTL1 and FTL2 are expressed outside the shoot apex and accumulate to much higher levels in plants grown in LDs than in SDs (Figures 1 and 3); fourth, both FTL1 and FTL2 were predicted to miR5200 targets and their mRNA could be indeed cleaved by miR5200 in vivo (Figure 1); fifth, the expression of FTL1 and FTL2 was severely reduced in miR5200 overexpression transgenic plants (Figure 2); and sixth, both FTL1 and FTL2 transcripts accumulated to higher levels in MIM5200 than in the wild-type plants under SDs (Figure 6). Nevertheless, differences between the two genes may exist. For instance, compared with FTL2, the base-pairing between miR5200 and FTL1 target site was not perfect, and most FTL1 cleavage events did not occur in the canonical miRNA cleavage site (Figure 1C), implying that miR5200 may mediate FTL1 translation repression by a mechanism other than mRNA degradation. Further isolation of single and double loss-of-function mutants is necessary to dissect FTL1 and FTL2 functions in the B. distachyon flowering pathway.
The transition from vegetative to reproductive stage is determined by daylength in many plant varieties. In Arabidopsis, CO is considered to be the central upstream regulator of FT expression in the photoperiod flowering pathway (Turck et al., 2008). Under LDs, both CO mRNA and protein are highly accumulated, thus resulting in activation of FT mRNA production (Kobayashi and Weigel, 2007). In SDs, there is very little FT expression because CO protein is largely degraded in a proteasome-dependent manner (Valverde et al., 2004). In Pooideae plants, the newly evolved miR5200 could also directly regulate FT transcripts. miR5200 accumulates abundantly to repress FT mRNAs in SD conditions, whereas miR5200 must, conversely, be diminished for the induction of FT in LDs (Figures 3 and 4). The different amounts of miR5200 with the daylength are partially due to the distinct chromatin conformations at pri-miRNA genes, as we observed that the status of positive and negative histone marks at MIR5200a and MIR5200b loci displayed profound differences under LDs and SDs (Figure 5).
Although the MIM5200 transgenic plants have a significant effect on flowering time in SDs, they do not flower extremely early like an LD plant. This is unlikely to be caused by the flowering time variation in independent transgenic events because the T1 segregated negative plants did not show any early-heading performance compared with the wild type. Moreover, the MIM5200 plants did not display any early-flowering phenotypes under LD conditions (see Supplemental Figure 9 online). These results convincingly suggest that miR5200 plays a role in the photoperiod-mediated flowering regulation. However, we propose the possibility that the target mimic strategy may not completely inhibit the activities of miR5200 in B. distachyon. Further genetic studies to find knockout miR5200 plants are essential for clearer understanding of miR5200 behaviors in flowering control. Another possibility is that additional factors other than miR5200 are also involved in photoperiod-mediated floral induction. This is almost certain, since the dynamic changes of FTL1 and FTL2 are not absolutely correlated with miR5200 alteration in the shifting experiments (Figure 4). We thereby propose a simple working model of the flowering pathway in Pooideae plants involving miR5200 (Figure 7B). It has been documented in wheat and barley that a pseudoresponse regulator protein named PPD1 functions upstream of CO and is critical in the response to photoperiodic flowering control (Turner et al., 2005; Shaw et al., 2012). Whether Pooideae miR5200 acts downstream or functions in parallel with PPD1 and CO-like proteins in the photoperiod pathway remains to be deciphered.
In all, our findings on miRNA-guided posttranscriptional modulation of florigen gene contribute insights into the precise regulatory modes of flowering in plants. Since most temperate cereals, including barley and wheat, belong to the Pooideae subfamily, artificial manipulation of miR5200 expression is perhaps a promising choice in crop breeding for proper heading date.
METHODS
Plant Materials and Growth Conditions
For most experiments, the Brachypodium distachyon accession Bd21 was used. B. distachyon and other Pooideae grasses were grown under SDs (8 h light/16 h dark) or LDs (16 h light/8 h dark) in growth chambers with temperatures of 22°C during the day and 16°C at night. Three- to four-week-old Bd21 seedlings harvested at Zeitgeber time 2 were used in the experiments, unless stated otherwise.
Constructs and Plant Transformation
The 517-bp transcription unit MIR5200a sequence and FTLs coding sequences were amplified from B. distachyon cDNA. FTLm was engineered by PCR-based mutation of miR5200 target site of FTL gene, and the miR5200 target mimic sequence was amplified from Arabidopsis thaliana and artificially modified as described before (Todesco et al., 2010). All these genes and target mimic constructs were under the control of maize (Zea mays) UBIQUITIN promoter for constitutive expression in B. distachyon plants. The sequences of primers for B. distachyon FT gene cDNA, miR5200 precursor amplification, and target mimic constructs are listed in Supplemental Table 1 online. Agrobacterium tumefaciens–mediated transformation of B. distachyon was essentially as previously described (Alves et al., 2009).
RNA Expression Analysis
Total RNA was extracted with Trizol reagent (Invitrogen) from three pooled B. distachyon plants grown under the indicated conditions. Thirty micrograms of enriched sRNAs were used for RNA gel blot detection of miR5200 amounts. 32P-end-labeled oligonucleotides complementary to miRNA sequences were used as probes. Total RNA was pretreated with RNase-free DNase I (Promega) to remove DNA and then reverse transcribed by EasyScript reverse transcriptase (TransGen Biotech) with oligo(dT) primers. RT-PCR and qPCR analyses of gene transcripts were performed as previously reported (Wu et al., 2010). qPCR was performed with SYBR Premix EX Taq (Takara). GAPDH mRNA was detected in parallel and used for data normalization. RT-PCR analyses for pri-miR5200, target gene expressions, and RNA gel blots for mature miR5200 were performed at least two times with similar results. qRT-PCRs were performed from three biological replicates with three technique repeats. Sequences of all probes and primers are listed in Supplemental Table 1 online.
RACE Mapping of MIRNA Primary Transcripts and Target Cleavage Site
5′ RNA ligase-mediated RACE reactions for pri-miR5200 transcript 5′-end determination were performed using poly(A)-enriched RNA that was pretreated with calf intestine phosphatase plus tobacco acid pyrophosphatase, and the cDNA was synthesized using random oligonucleotides as primers (Ambion). For 3′ MIRNA primary transcripts, cDNA was synthesized using adaptor-tagged oligo(dT) primers. 5′ RACE of miR5200 target sites of FT genes was performed using poly(A) -enriched RNA without calf intestine phosphatase plus tobacco acid pyrophosphatase treatment. All amplification products were gel purified and cloned in the pGEM-T easy vector (Promega) for sequencing.
DNA Methylation and Histone Modification Analyses
Genomic DNA was isolated from 3-week-old B. distachyon seedlings grown in SDs and LDs using the DNA secure plant kit (Tiangen). Bisulfite sequencing and PCR-based DNA methylation assays were conducted as described previously (Wu et al., 2010). The PCR products were cloned into the pGEM-T easy vector (Promega), and at least 20 individual clones were sequenced. ChIP was performed as described before with minor modifications. Two grams of 3-week-old B. distachyon tissue was treated with formaldehyde buffer for 20 min at room temperature for cross-linking by vacuum infiltration, followed by addition of 125 mM Gly. Plants were rinsed with water, frozen, and ground into powder in liquid nitrogen using a mortar and pestle, suspended in 30 mL of Extraction Buffer I (10 mM Tris-HCl, pH 8.0, 0.5 M Suc, 10 mM MgCl2, 5 mM DTT, 1 mM PMSF, and 1% Roche protease inhibitors), and filtered through two layers of Miracloth and centrifuged at 3000g at 4°C for 15 min to remove debris. Nuclear pellets were resuspended in 1 mL of Extraction Buffer II (10 mM Tris-HCl, pH 8.0, 0.5 M Suc, 10 mM MgCl2, 0.5% Triton X-100, 5 mM DTT, 1 mM PMSF, and 1% tablet Roche protease inhibitors) and centrifuged at 12,000g at 4°C for 10 min. Then the pellets were suspended in nuclei lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS, 5 mM DTT, and 1% Roche protease inhibitors) and sonicated five times for 20 s each. After centrifugation at 16,000g for 10 min, the supernatant was diluted 10-fold with ChIP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, and 167 mM NaCl). After being precleared for 1 h, 25 μL of protein A agarose (Roche) and the appropriate antibodies were added for incubation overnight. Agarose-antibody complexes were washed five times, 5 min each, with low-salt, high-salt, LiCl washing buffer, and TE buffer. Elution and reverse cross-linking were performed as described previously (Saleh et al., 2008). One microliter of extracted DNA was used as template to amplify the indicated sequences for histone modification status.
Sequence Alignment and Phylogenetic Analysis
BLASTP was performed using the deduced amino acid sequences of FT genes from B. distachyon, Arabidopsis thaliana, and barley (Hordeum vulgare). Multiple sequence alignment was performed using ClustalW, and the construction of phylogenetic tree was conducted by MEGA program using the neighbor-joining method. Bootstrap analysis was performed to estimate nodal support on the basis of 1000 replicates.
Accession Numbers
Sequence data from this article can be found in the Brachypodium Genome Initiative, Arabidopsis Genome Initiative, or GenBank databases under the following accession numbers: FTL1, Bradi2g07070; FTL2, Bradi1g48830; FTL4, Bradi4g35040; FTL5, Bradi5g14010; FTL6, Bradi3g48036; FTL7a, Bradi4g39730; FTL7b, Bradi4g39750; FTL7c, Bradi4g39760; FTL9, Bradi2g49795; FTL10, Bradi2g19670; FTL12, Bradi1g38150; FTL13, Bradi3g08890; VRN1, Bradi3g08340; FUL2, Bradi1g59250; DCL1, Bradi1g77090; MIR5200a (1-14), KF638635-638648; MIR5200b, KF638649; Arabidopsis FT, At1g15480; Arabidopsis TSF, At4g20370; barley FT1, DQ100327; barley FT2, DQ297407; barley FT3, DQ411319; barley FT4, DQ411320; and barley FT5, EF012202.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Flowering Habit of Brachypodium Accession Bd21.
Supplemental Figure 2. Conservation of miR5200 in Pooideae Plants.
Supplemental Figure 3. Gene Structures of Pri-miR5200a and Pri-miR5200b.
Supplemental Figure 4. FT-Like Orthologous Genes Are Potential miR5200 Targets in Wheat and Barley.
Supplemental Figure 5. Effects of miR5200 Ectopic Expression on Flowering Time and Plant Height.
Supplemental Figure 6. Transcriptional Control of miR5200 by Photoperiod.
Supplemental Figure 7. DNA Methylation Analysis at Pri-miR5200 Loci in Brachypodium Grown under SDs and LDs.
Supplemental Figure 8. Extremely Early Flowering Phenotypes of Transgenic Plants Ectopically Expressing Either FTL1 or FTL2 Gene with or without miR5200 Target Sites.
Supplemental Figure 9. miR5200 Target Mimicry Specifically Alters Flowering Time in Transgenic Plants under SD Conditions.
Supplemental Table 1. Oligos Used as Primers and Probes in This Study.
Supplemental Data Set 1. Text File of Alignment Corresponding to Phylogenetic Analysis in Supplemental Figure 4.
Acknowledgments
We thank Lihui Li, Yuqing Lu, and Lihong Miao for providing the seeds of Pooideae grasses. We also thank Yijun Qi, Jiawei Wang, and Detlef Weigel for critical reading of the article. This work was supported in part by the National Natural Science Foundation of China (31200181 and 30871323), the National Basic Research “973” (2009CB118306 and 2010CB125902), and a Core Research Budget of the Non-profit Governmental Research Institution (Institute of Crop Science, Chinese Academy of Agricultural Sciences).
AUTHOR CONTRIBUTIONS
L.W. and L.M. designed the research. L.W., Do.L., J.W., Z.Q., R.Z., and Da.L. performed the experiments. L.W., Do.L., A.L., D.F., W.Z., and L.M. analyzed the data. L.W. and L.M. wrote the article. L.W., W.Z., and L.M. contributed equally to this work.
Glossary
- LD
long-day SD: short-day
- miRNA
microRNA
- sRNA
small RNA
- qPCR
quantitative real-time PCR
- ChIP
chromatin immunoprecipitation
- RACE
rapid amplification of cDNA ends
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Abrouk M., Zhang R., Murat F., Li A., Pont C., Mao L., Salse J. (2012). Grass microRNA gene paleohistory unveils new insights into gene dosage balance in subgenome partitioning after whole-genome duplication. Plant Cell 24: 1776–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian J., Farrona S., Reimer J.J., Albani M.C., Coupland G., Turck F. (2010). cis-regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves S.C., Worland B., Thole V., Snape J.W., Bevan M.W., Vain P. (2009). A protocol for Agrobacterium-mediated transformation of Brachypodium distachyon community standard line Bd21. Nat. Protoc. 4: 638–649 [DOI] [PubMed] [Google Scholar]
- Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Bäurle I., Dean C. (2006). The timing of developmental transitions in plants. Cell 125: 655–664 [DOI] [PubMed] [Google Scholar]
- Bergonzi S., Albani M.C., Ver Loren van Themaat E., Nordström K.J., Wang R., Schneeberger K., Moerland P.D., Coupland G. (2013). Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340: 1094–1097 [DOI] [PubMed] [Google Scholar]
- Brkljacic J., et al. (2011). Brachypodium as a model for the grasses: Today and the future. Plant Physiol. 157: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C., Pelaz S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Candela H., Hake S. (2009). Big impacts by small RNAs in plant development. Curr. Opin. Plant Biol. 12: 81–86 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A., Li C., Dubcovsky J. (2009). Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 12: 178–184 [DOI] [PubMed] [Google Scholar]
- Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., Yano M., Yoshimura A. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S., Coupland G., Turck F. (2008). The impact of chromatin regulation on the floral transition. Semin. Cell Dev. Biol. 19: 560–573 [DOI] [PubMed] [Google Scholar]
- Filiz E., Ozdemir B.S., Budak F., Vogel J.P., Tuna M., Budak H. (2009). Molecular, morphological, and cytological analysis of diverse Brachypodium distachyon inbred lines. Genome 52: 876–890 [DOI] [PubMed] [Google Scholar]
- Fornara F., de Montaigu A., Coupland G. (2010). SnapShot: Control of flowering in Arabidopsis. Cell 141: 550, e1–e2 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Hackenberg M., Shi B.J., Gustafson P., Langridge P. (2012). A transgenic transcription factor (TaDREB3) in barley affects the expression of microRNAs and other small non-coding RNAs. PLoS ONE 7: e42030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.A., Bailey P.C., Laurie D.A. (2010). Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE 5: e10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.Y., Lee S., Seo P.J., Yang M.S., Park C.M. (2010). Identification and molecular characterization of a Brachypodium distachyon GIGANTEA gene: Functional conservation in monocot and dicot plants. Plant Mol. Biol. 72: 485–497 [DOI] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kim J.J., Lee J.H., Kim W., Jung H.S., Huijser P., Ahn J.H. (2012). The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 159: 461–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Weigel D. (2007). Move on up, it’s time for change—Mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21: 2371–2384 [DOI] [PubMed] [Google Scholar]
- Komiya R., Yokoi S., Shimamoto K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136: 3443–3450 [DOI] [PubMed] [Google Scholar]
- Li C., Dubcovsky J. (2008). Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 55: 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C., Shen L., Wu Y., Chen H., Robertson M., Helliwell C.A., Ito T., Meyerowitz E., Yu H. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas S.J., Budak H. (2012). Sorting the wheat from the chaff: Identifying miRNAs in genomic survey sequences of Triticum aestivum chromosome 1AL. PLoS ONE 7: e40859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P.A., Koenig D., Weigel D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 109: 2461–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Muszynski M.G., Danilevskaya O.N. (2011). The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23: 942–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C., Abelenda J.A., Cruz-Oró E., Cuéllar C.A., Tamaki S., Silva J., Shimamoto K., Prat S. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478: 119–122 [DOI] [PubMed] [Google Scholar]
- Opanowicz M., Vain P., Draper J., Parker D., Doonan J.H. (2008). Brachypodium distachyon: Making hay with a wild grass. Trends Plant Sci. 13: 172–177 [DOI] [PubMed] [Google Scholar]
- Pin P.A., Benlloch R., Bonnet D., Wremerth-Weich E., Kraft T., Gielen J.J., Nilsson O. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330: 1397–1400 [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Sawa M., Kay S.A. (2011). GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 11698–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A.W., Shi B.J., Huang C.Y., Langridge P., Baumann U. (2011). Discovery of barley miRNAs through deep sequencing of short reads. BMC Genomics 12: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C.J., Doyle M.R., Manzaneda A.J., Rey P.J., Mitchell-Olds T., Amasino R.M. (2010). Natural variation of flowering time and vernalization responsiveness in Brachypodium distachyon. Bioenerg. Res. 3: 38–46 [Google Scholar]
- Seo E., Yu J., Ryu K.H., Lee M.M., Lee I. (2011). WEREWOLF, a regulator of root hair pattern formation, controls flowering time through the regulation of FT mRNA stability. Plant Physiol. 156: 1867–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L.M., Turner A.S., Laurie D.A. (2012). The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum). Plant J. 71: 71–84 [DOI] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti S., Fornara F., Vincent C., Andrés F., Nordström K., Göbel U., Knoll D., Schoof H., Coupland G. (2012). Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24: 444–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B., Bagnall D.J., Ellis M.H., Peacock W.J., Dennis E.S. (2003). MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. USA 100: 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F., Fornara F., Coupland G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R.P., Laurie D.A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Vogel J.P., et al. International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). MiR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wei B., Cai T., Zhang R., Li A., Huo N., Li S., Gu Y.Q., Vogel J., Jia J., Qi Y., Mao L. (2009). Novel microRNAs uncovered by deep sequencing of small RNA transcriptomes in bread wheat (Triticum aestivum L.) and Brachypodium distachyon (L.) Beauv. Funct. Integr. Genomics 9: 499–511 [DOI] [PubMed] [Google Scholar]
- Wenkel S., Turck F., Singer K., Gissot L., Le Gourrierec J., Samach A., Coupland G. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A. (2011). FT, a mobile developmental signal in plants. Curr. Biol. 21: R374–R378 [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009a). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.J., Ma Y.K., Chen T., Wang M., Wang X.J. (2012). PsRobot: A web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 40 (Web Server issue): W22–W28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zhang Q., Zhou H., Ni F., Wu X., Qi Y. (2009b). Rice microRNA effector complexes and targets. Plant Cell 21: 3421–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zhou H., Zhang Q., Zhang J., Ni F., Liu C., Qi Y. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell 38: 465–475 [DOI] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Fu D., Li C., Blechl A., Tranquilli G., Bonafede M., Sanchez A., Valarik M., Yasuda S., Dubcovsky J. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Xu Y., Huan Q., Chong K. (2009). Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics 10: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C.M., Zhang T.Q., Wang X., Yu S., Lian H., Tang H., Feng Z.Y., Zozomova-Lihová J., Wang J.W. (2013). Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science 340: 1097–1100 [DOI] [PubMed] [Google Scholar]