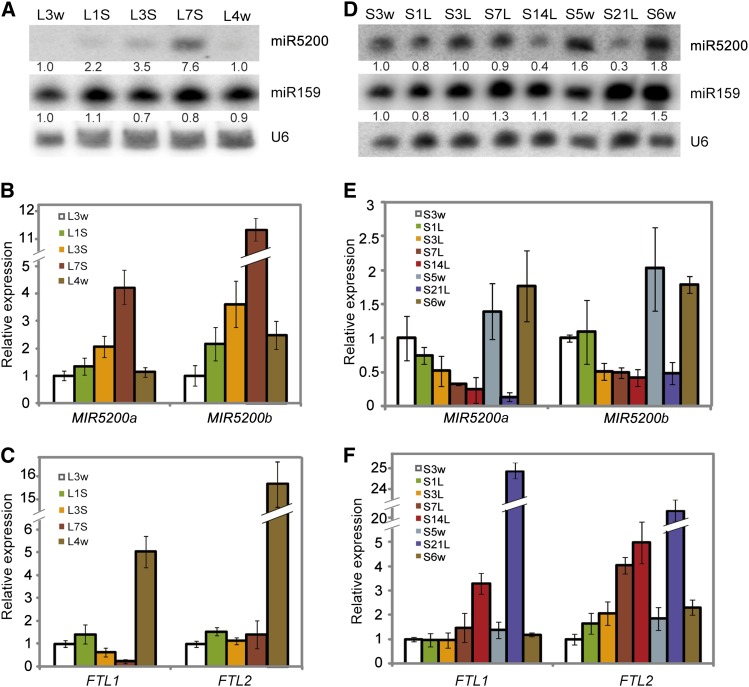
Figure 4.
Dynamic Effects of Daylength Changes on miR5200, FTL1, and FTL2 Accumulation.
(A) Time course of miR5200 expression when B. distachyon plants were shifted from LDs to SDs. L3w and L4w indicate that B. distachyon were grown under LDs for 3 and 4 weeks. L1S, L3S, and L7S represent 3-week-old plants that were moved from LDs to SDs for 1, 3, and 7 d, respectively. Numbers below each miRNA gel blot indicate fold changes relative to the miRNA level in wild-type Bd21 plants grown under LDs for 3 weeks.
(B) qPCR examination of dynamic changes of MIR5200a and MIR5200b in (A).
(C) qPCR analysis of dynamic effects of daylength on FTL1 and FTL2 in (A).
(D) Time course of miR5200 expression when 3-week-old SD plants were transferred to LDs for 1, 3, 7, 14, and 21 d. Plants grown under SDs for 5 and 6 weeks were harvested and used as controls. Numbers below each miRNA gel blot denote fold changes relative to the miRNA level in Bd21 grown under SDs for 3 weeks.
(E) qPCR examination of dynamic changes of MIR5200a and MIR5200b in (D).
(F) qPCR analysis of dynamic effects of daylength on FTL1 and FTL2 in (D). Error bar means sd (n = 3). Samples for RNA extraction in the experiment were collected at Zeitgeber time 2. U6 was used as loading control for RNA gel blots. GAPDH was used as an internal control for normalization of qPCR data.