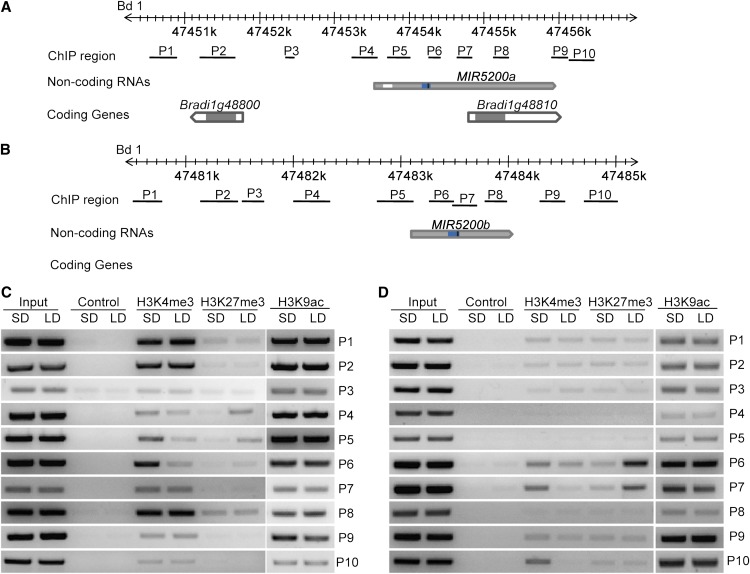
Figure 5.
Histone Modification Status at Pri-miR5200 Loci in B. distachyon Grown under Different Daylengths.
(A) and (B) Diagrams showing the genomic locations, the adjacent coding genes, and the regions examined by ChIP assay for MIR5200a (A) and MIR5200b (B). Gray boxes represent exons, and white boxes represent introns. Blue boxes located in gray boxes indicate predicted MIR5200a and MIR5200b hairpin structures. Positions of mature miR5200 are set as black bars. ChIP regions are shown by black lines.
(C) and (D) Relative abundance of H3K4me3, H3K27me3, and H3K9ac at MIR5200a (C) and MIR5200b (D) genomic regions in B. distachyon grown under SDs and LDs. Note that the high levels of H3K4me3 and the decreased H3K27me3 around regions corresponding to MIR5200a (P4/P5/P6) and MIR5200b (P6/P7) hairpin structures are associated with the activation of MIR5200a and MIR5200b expression under SD conditions.