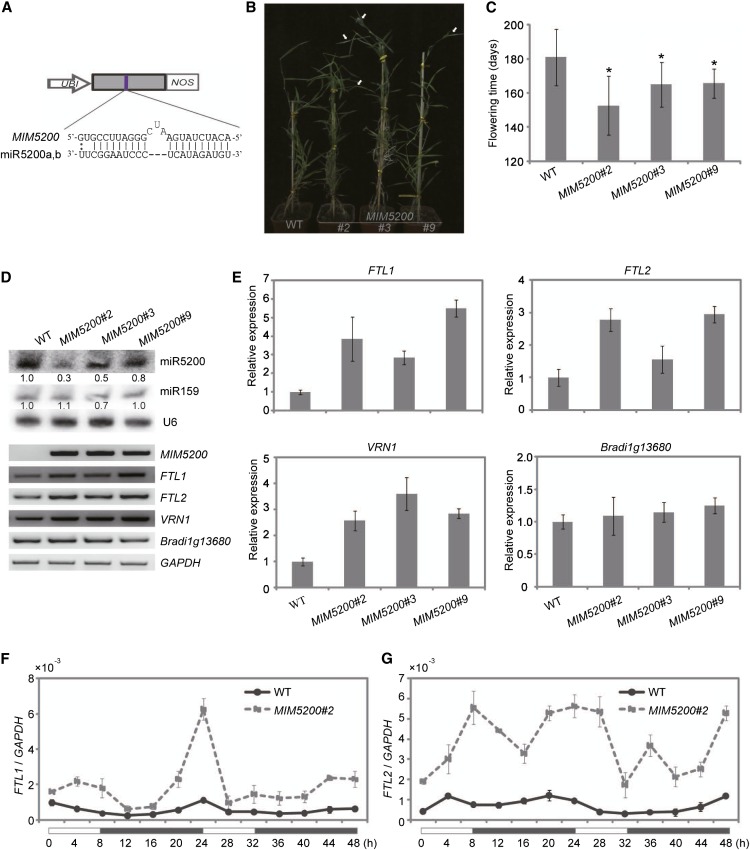
Figure 6.
Disruption of miR5200 Activity in B. distachyon Promotes Flowering under SDs.
(A) A diagram of MIM5200 showing the designed sequences for miR5200 target mimics. UBI, maize ubiquitin promoter; NOS, nos terminator.
(B) Representative photograph of flowering phenotypes in 6-month-old wild-type (WT) and indicated T1 MIM5200 transgenic plants grown in SDs. The white arrows point to spikes.
(C) Flowering time of wild-type and the indicated T1 MIM5200 transgenic lines grown in SDs. Error bar shows sd (n = 8). Asterisks indicate a significant difference between wild-type and transgenic plants (Student’s t test, P value < 0.05).
(D) Expression of miR5200 (by RNA gel blots), targets, and other flowering genes (by RT-PCR) as well as Bradi1g13680 (miR169 target gene as a control) in wild-type and the indicated T1 MIM5200 transgenic lines under SDs. U6 and GAPDH were served as the RNA gel blot and RT-PCR RNA loading controls, respectively. The numbers below each miRNA gel blot denote the fold changes relative to the miRNA level in the wild type.
(E) qPCR analysis of miR5200 target genes, VRN1, and Bradi1g13680 (miR169 target as a control) expression in wild-type and the indicated MIM5200 transgenic plants under SDs.
(F) and (G) Diurnal expression patterns of FTL1 (F) and FTL2 (G) in wild-type and the indicated MIM5200 transgenic plants under SDs. Each point represents the average of three technique replicates, and the error bar indicates sd. White and black bars along the horizontal axes represent light and dark periods, respectively. Numbers below the horizontal axes indicate the time in hours.