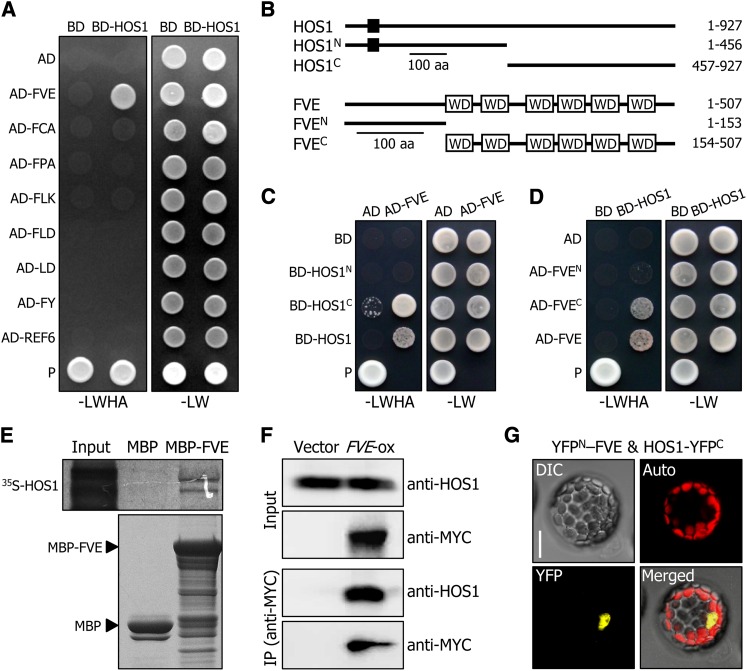
Figure 3.
HOS1 Interacts with FVE in the Nucleus.
(A) Interactions of HOS1 with FVE in yeast cells. Autonomous flowering pathway genes and the HOS1 gene were fused in frame to the GAL4 activation domain (AD)–coding and GAL4 DNA binding domain (BD)–coding sequences, respectively. The HOS1–FVE interactions were assayed by cell growth on selective medium. –LWHA indicates Leu, Trp, His, and Ade dropout plates. –LW indicates Leu and Trp dropout plates. P, positive control.
(B) Deletion constructs of HOS1 and FVE. Numbers indicate residue positions. Black box, REALLY INTERESTING NEW GENE finger domain. WD, WD40 domain. aa, amino acids.
(C) and (D) Interactions of HOS1 with FVE in yeast cells. The HOS1 (C) or FVE (D) constructs were fused with GAL4 BD or GAL4 AD, respectively. Yeast two-hybrid assays were performed as described in (A).
(E) In vitro pull-down assays. Recombinant MBP-FVE fusion and in vitro–translated 35S-labeled HOS1 polypeptides were used (top panel). Part of a Coomassie blue–stained gel is shown (bottom panel). The input represents 5% of the radiolabeled protein used in the assays.
(F) Coimmunoprecipitation assays. Total proteins were extracted from transgenic plants overexpressing a FVE-MYC gene fusion (FVE-ox) grown on MS-agar plates for 6 d. Protein complexes were immunoprecipitated using an anti-MYC antibody and detected immunologically using an anti-MYC or anti-HOS1 antibody. Transgenic plants harboring the 6xMYC-pBA vector alone were used as control plants.
(G) BiFC assays. The YFPN-FVE and HOS1-YFPC constructs were coexpressed transiently in Arabidopsis protoplasts and visualized by differential interference contrast microscopy and fluorescence microscopy. Bar = 20 μm.