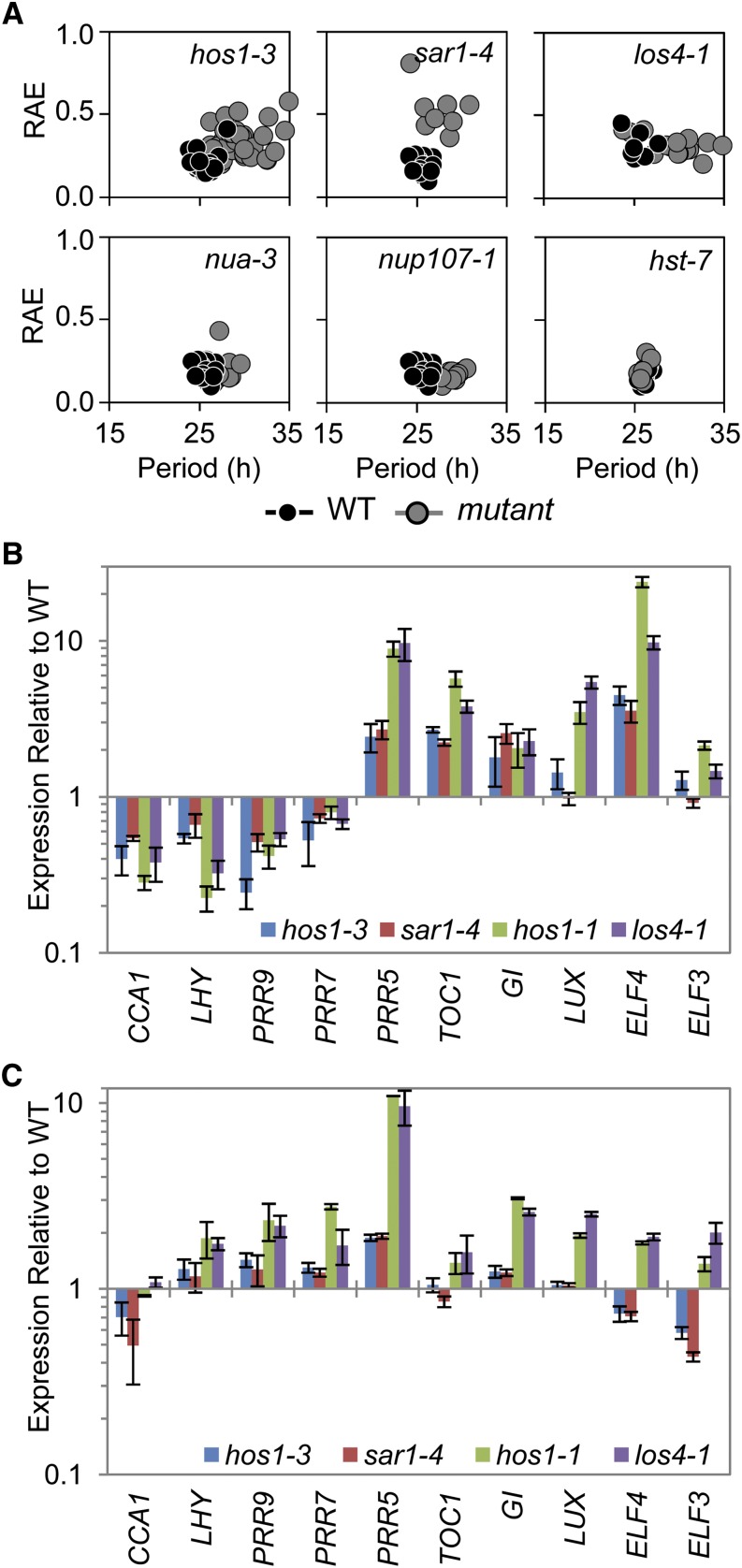
Figure 6.
Mutants Resulting in Altered mRNA Export Exhibit Long-Period Circadian Phenotypes and Alterations to Clock Gene Expression Similar to hos1.
(A) Delayed fluorescence was performed on a range of nuclear pore–associated mutants. Seedlings were grown in 12L:12D white-light cycles for 16 d prior to transferring to constant 17°C and constant red light for analysis. Each plot shows period estimations plotted versus RAE comparing the wild type (WT; black circles) and mutant (gray circles).
(B) and (C) RNA analysis comparing the expression of clock genes in hos1 alleles, sar1-4 and los4-1, to their appropriate wild types. Plants were grown for 2 weeks in 12L:12D white-light cycles and harvested in triplicate at dawn CT0 (B) or dusk CT12 (C). Data are shown relative to the wild-type values for comparison. Data are averages of biological triplicate ± se normalized for transcript abundance to TUB9 (B) or UBQ10 (C). Bar colors are as follows: hos1-3, blue; sar1-4, red; hos1-1, green; and los4-1, purple.