Figure 3. Expression and cellular origin of GOLM1-MAK10 in ESCC patient tissues and cancer cell lines.
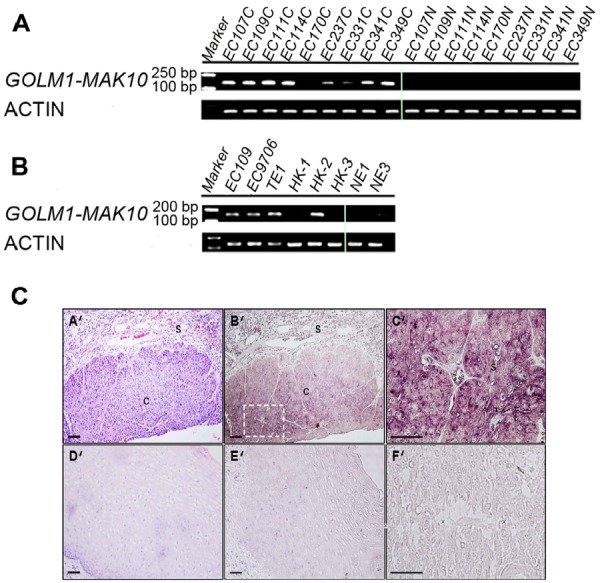
(A) GOLM1-MAK10 was readily detectable by RT-PCR, and differentially expressed in cancer vs. matched benign tissue. The expression of chimeric RNA in 9 patients are shown. Left side: cancer tissues. Right side: matched benign tissues. (B) GOLM1-MAK10 was expressed in some ESCC cell lines, including EC109, EC9706, TE1, HK1, HK2, and HK3, but minimally expressed in immortalized esophageal epithelial cells NE1 and NE3. Both (A) and (B) suggest that GOLM1-MAK10 is enriched in cancer cells. (C) Cellular origin of GOLM1-MAK10 RNA in tissue sections of ESCC patient as determined by in situ hybridization. (A'), (B'), (C'), and (F'): cancerous esophageal epithelium. (D') and (E'): non-neoplastic esophageal epithelium. Hematoxylin/eosin staining identifies cancer mass with intense purple color in cancerous esophageal epithelium (A'), which is largely absent in non-neoplastic esophageal epithelium (D'). Hybridization with antisense RNA probe reveals the location of chimeric RNAs with dark maroon color in cancerous esophageal epithelium (B'). Note that the areas positive for chimeric RNAs in B' co-localize with cancer mass in AB'. Stromal cells surrounding tumor tissue also showed slight elevated staining but to a lesser extent. In contrast, non-neoplastic esophageal epithelium shows little or no staining with antisense probe (E'). The inset in B' is magnified as C', which reveals the cytoplasmic localization of chimeric RNAs in cancer cells. F' is an adjacent section of C' stained with sense probe as control, which resulted in little signal. s: stromal; c: cancer. Scale bar= 100 um.
