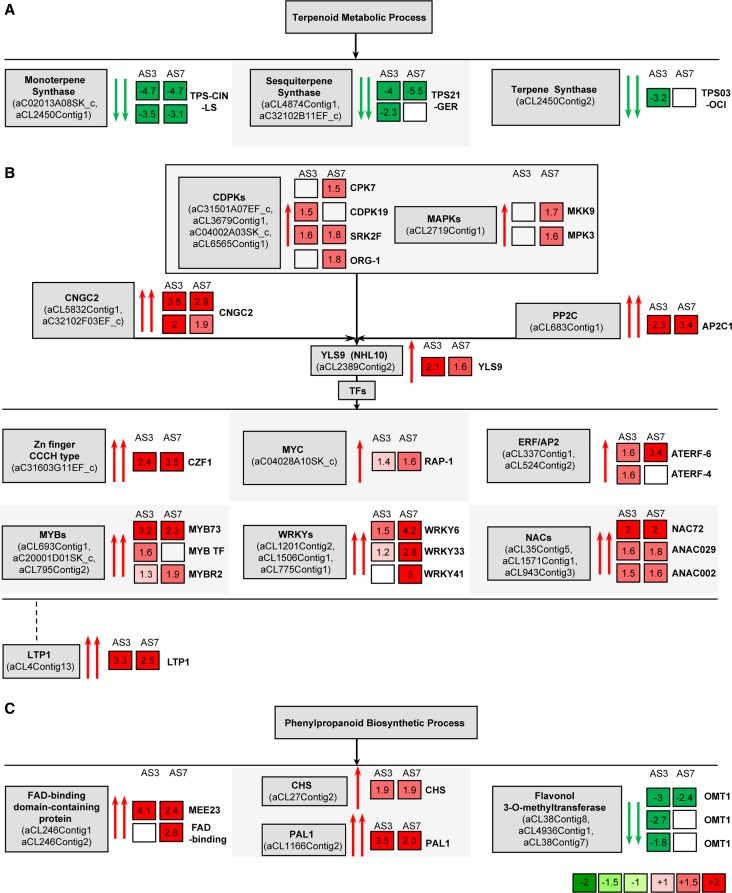
Figure 5.
Graphical representation of the genes that were differentially expressed in the flavedo of intact oranges. A, Genes involved in the terpenoid metabolic process that were down-regulated in transgenic orange plants. B, Defense-related genes that were up-regulated in transgenic orange plants. C, Genes involved in the phenylpropanoid biosynthetic process that were misregulated in transgenic orange plants. Numbers in squares indicate the ratio of expression in AS fruits compared with EV fruits. Blank squares indicate genes not detected. The most similar Arabidopsis gene functions are listed at the right side of the squares. Genes are as follows: TPS-CIN-LS, monoterpene synthase similar to limonene synthase; TPS21-GER, sesquiterpene synthase similar to germacrene-d synthase; TPS03-OCI, terpene synthase similar to β-ocimene/α-farnesene synthase; CDPKs, CPK7, CDPK19, SRK2F, and ORG-1; MAPKs, MKK9 and MPK3; CNGC2, cyclic nucleotide-regulated ion channel; PP2C, protein phosphatase 2C (AP2C1); YLS9; Zn (zinc) finger CCCH type family protein, CZF1; MYC, basic helix-loop-helix protein RAP-1; ERF/AP2, ethylene response factor/AP2 domains (ATERF6 and ATERF4); MYB, members of the MYB family TF (MYB73, MYB TF, and MYBR2); WRKY, members of the WRKY family TF (WRKY6, WRKY33, and WRKY41); NAC, NAC72, ANAC029, and ANAC002; LTP1; FAD, FAD-binding proteins (MEE23 and FAD-binding); CHS, chalcone synthase; PAL1; and flavonol 3-O-methyltransferase (OMT1).