Rice utilizes different type of Rubisco small subunit in nonphotosynthetic cells.
Abstract
Rubisco small subunits (RbcSs) are encoded by a nuclear multigene family in plants. Five RbcS genes, OsRbcS1, OsRbcS2, OsRbcS3, OsRbcS4, and OsRbcS5, have been identified in rice (Oryza sativa). Among them, the amino acid sequence of OsRbcS1 differs notably from those of other rice RbcSs. Phylogenetic analysis showed that OsRbcS1 is genetically distant from other rice RbcS genes and more closely related to RbcS from a fern and two woody plants. Reverse transcription-PCR and promoter β-glucuronidase analyses revealed that OsRbcS1 was not expressed in leaf blade, a major photosynthetic organ in rice, but was expressed in leaf sheath, culm, anther, and root central cylinder. In leaf blade of transgenic rice overexpressing OsRbcS1 and leaf sheath of nontransgenic rice, OsRbcS1 was incorporated into the Rubisco holoenzyme. Incorporation of OsRbcS1 into Rubisco increased the catalytic turnover rate and Km for CO2 of the enzyme and slightly decreased the specificity for CO2, indicating that the catalytic properties were shifted to those of a high-activity type Rubisco. The CO2 assimilation rate at low CO2 partial pressure was decreased in overexpression lines but was not changed under ambient and high CO2 partial pressure compared with nontransgenic rice. Although the Rubisco content was increased, Rubisco activation state was decreased in overexpression lines. These results indicate that the catalytic properties of Rubisco can be altered by ectopic expression of OsRbcS1, with substantial effects on photosynthetic performance in rice. We believe this is the first demonstration of organ-specific expression of individual members of the RbcS gene family resulting in marked effects on Rubisco catalytic activity.
Rubisco is the key enzyme catalyzing the first step of photosynthetic CO2 assimilation by carboxylation of ribulose-1,5-bisphosphate (RuBP). Rubisco also catalyzes the competing oxygenation of RuBP, leading to wasteful photorespiration. Rubisco is considered to be a rate-limiting enzyme of C3 photosynthesis under the conditions of saturated light intensity and present ambient CO2 concentration due to its extremely slow catalytic turnover rate (kcat) and its role in photorespiration (von Caemmerer and Quick, 2000). To compensate for these inefficient enzymatic properties, C3 plants accumulate a large amount of Rubisco, accounting for 15% to 30% of total leaf nitrogen, making Rubisco catalytic efficiency a key parameter underpinning leaf level nitrogen use efficiency (Evans, 1989; Makino et al., 1992). Also, these observations make Rubisco a potential target for the improvement of photosynthesis and agricultural productivity (Peterhansel and Offermann, 2012; Parry et al., 2013).
In most chemoautotrophic bacteria, cyanobacteria, algae, and all land plants, Rubisco holoenzyme comprises eight large subunits (RbcLs) and eight small subunits (RbcSs), i.e. L8S8 structure (Andersson and Backlund, 2008). RbcL is encoded by a single gene on the multicopy chloroplast DNA, while RbcS is encoded by a multigene family in the nucleus. There is substantial natural variation in the kinetic properties of L8S8 Rubisco even in vascular plants. C4 plants with a CO2 concentrating mechanism and C3 plants from cool habitats, where photorespiration is reduced, contain Rubisco with a high kcat (Sage, 2002). According to structural-functional analyses, most of the important amino acid residues necessary for catalysis are present in RbcL. In accordance with this, the kinetic properties of Rubisco in an interspecific hybrid will follow maternal inheritance (Evans and Austin, 1986). In addition, Whitney et al. (2011) reported that hybrid Rubisco composed of Flaveria bidentis C4 RbcL and tobacco (Nicotiana tabacum) RbcS showed the catalytic properties of high-activity-type Rubisco from C4 Flaveria bidentis. These results suggest that the catalytic properties of Rubisco are largely determined by the amino acid sequence of RbcL. On the other hand, it was reported that the kinetic performance of a hybrid Rubisco composed of Chlamydomonas reinhardtii RbcL and vascular plant RbcS partially shifted to that of vascular plants (Karkehabadi et al., 2005; Genkov et al., 2010). In previous work, we clearly showed that chimeric incorporation of RbcS from the C4 plant sorghum (Sorghum bicolor) greatly enhanced the kcat of Rubisco in transgenic rice (Oryza sativa; Ishikawa et al., 2011). Hence, it is proposed that RbcS as well as RbcL can make a significant contribution to the catalytic performance of Rubisco across species. The sequences of RbcL are highly conserved among vascular plants, whereas those of RbcS are much more divergent (Spreitzer, 2003). Considering these observations, RbcS could actually be a key factor in determining interspecific variation in kinetic properties of Rubisco. In addition to its enzymatic functions, RbcS is also involved in quantitative regulation of Rubisco via coordinated expression of RbcL and RbcS in plants (Rodermel et al., 1996; Suzuki and Makino, 2012).
The number of genes comprising the RbcS multigene family varies depending on species, ranging from two in C. reinhardtii to 22 or more in wheat (Triticum aestivum; Spreitzer, 2003). Individual members of the RbcS gene family sometimes show different expression patterns in leaf development and tissue specificity (Dean et al., 1989; Silverthorne and Tobin, 1990); however, these expressions are essentially confined to photosynthetic tissues. In most plants, the amino acid sequences of RbcS share high identity with each other within a gene family and are often identical in mature protein (Spreitzer, 2003). Hence, it has been assumed that there are no functional differences in the RbcS gene family within a species. In rice, five RbcS genes, OsRbcS1, OsRbcS2, OsRbcS3, OsRbcS4, and OsRbcS5, have been identified (Suzuki et al., 2007). OsRbcS2 to OsRbcS5 are all located on chromosome 12 and arranged in a tandem array at a single locus. The deduced amino acid sequences of OsRbcS2 to OsRbcS5, without the transit peptide for targeting to the chloroplast, are completely identical. By contrast, OsRbcS1 is located on chromosome 2 and has remarkably lower homology with other rice RbcSs (55.4% similarity in mature protein; Suzuki et al., 2007). Furthermore, it has been reported that OsRbcS1 is not appreciably expressed in leaf blade, the major photosynthetic organ in rice, at any time in the plant’s lifecycle (Suzuki et al., 2009). Other than OsRbcS1, to our knowledge there are no reports, in any plants, of RbcS genes showing such a large diversity of amino acid sequences and expression patterns. Considering these distinct features, OsRbcS1 might be expected to have some specialized functions different from other members of the RbcS gene family.
In this study, we focus on this unusual RbcS in rice and report on expression analyses and functional analyses using transgenic rice. It was found that OsRbcS1 functions in encoding the small subunit of Rubisco in particular tissues such as leaf sheath, and the holoenzyme containing OsRbcS1 has altered kinetic properties similar to the high-catalytic-activity Rubisco typically found in C4 plants.
RESULTS
RbcS Gene Family in Rice
Deduced amino acid sequences of the coding region of the rice RbcS gene family were obtained from Rice Anotation Project Database (RAP-DB; http://rapdb.dna.affrc.go.jp) and aligned within the gene family or with RbcS from other species (Supplemental Figs. S1 and S2). OsRbcS2 to OsRbcS5 contain only one intron, and this interrupts the coding region between residues 49 to 50, a common characteristic of monocotyledonous RbcS, whereas OsRbcS1 has an additional intron interrupting the coding region between residues 96 to 97 as observed in many dicotyledonous RbcS (Supplemental Fig. S1; Dean et al., 1989). As reported by Suzuki et al. (2007), OsRbcS2 to OsRbcS5 are highly similar to one another at the amino acid level. The mature proteins of OsRbcS2 to OsRbcS5 without chloroplast-targeting transit peptides, are identical. By contrast, the mature protein of OsRbcS1 is notably different and shares only 55.4% similarity with the proteins encoded by other rice RbcSs. In particular, the regions of the N terminus of mature protein, the first half of the βA-βB loop and the C terminus in OsRbcS1 showed a relatively higher degree of divergence from those in other rice RbcSs (Supplemental Fig. S2). However, 10 highly conserved amino acid residues among RbcS reported by Spreitzer (2003) are mostly conserved in OsRbcS1, except for one apparently insignificant substitution of Tyr-32 by Phe in OsRbcS1. In addition, the important amino acid residue Arg-54, which forms a hydrogen bond with RbcL in the assembly domain of RbcS (Wasmann et al., 1989), is also conserved in OsRbcS1. Thus, it is anticipated that OsRbcS1 could encode a functional small subunit of Rubisco.
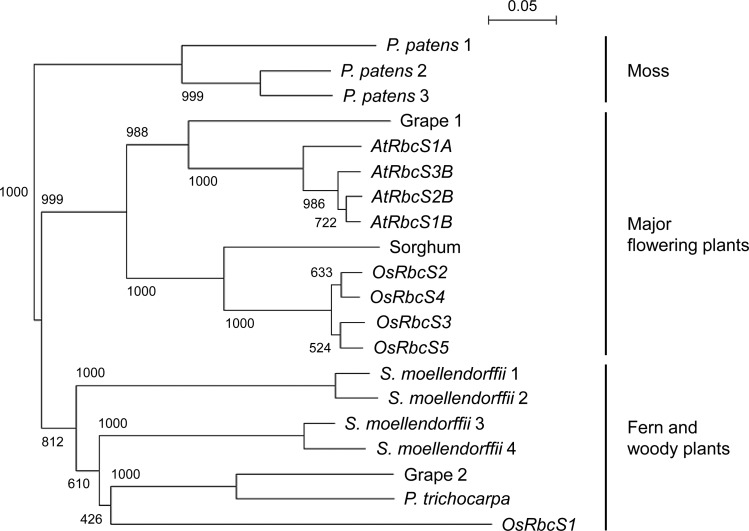
Phylogenetic analysis of RbcS from terrestrial plants revealed that the closely related OsRbcS2 to OsRbcS5 were classified into the group consisting of major flowering plants (Fig. 1). However, OsRbcS1 was distantly related to other rice RbcSs and classified into another group, consisting of RbcS from a fern and two woody plants. It was found that grape (Vitis vinifera) also contains two genetically distant types of RbcS, as in rice. In addition, we also found divergent OsRbcS1-like genes in the published genomes of some plant species such as foxtail millet (Setaria italica; accession no. XM_004954652) and tomato (Solanum lycopersicum; XM_004243031), suggesting that a wide variety of plants could contain genes orthologous to OsRbcS1. To study the distribution of OsRbcS1-like genes among the Poaceae, DNA gel-blot analysis was performed (Supplemental Fig. S3). DNA bands hybridized with OsRbcS1-specific probes could be detected in rice, wild rice species such as Oryza rufipogon (AA genome) and Oryza officinalis (CC genome), and foxtail millet, whereas no clear bands were observed in barley (Hordeum vulgare), maize (Zea mays), and rye (Secale cereale). These results indicate that the presence of OsRbcS1 ortholog is not always common, even among the Poaceae, and not necessarily related to evolutionary conservation.
Figure 1.
Phylogenetic tree of RbcS from terrestrial plants. The tree was constructed by the neighbor-joining method using the deduced amino acid sequences of the coding region of RbcS. The bootstrap values calculated as per mil for 1,000 replications are shown at nodes. The accession numbers are Physcomitrella patens 1, Pp059935; P. patens 2, Pp125903; P. patens 3, Pp115069; Arabidopsis AtRbcS1A, AT1G67090; AtRbcS1B, AT5G38430; AtRbcS2B, AT5G38420; AtRbcS3B, AT5G38410; sorghum, Sb05g003480; rice OsRbcS1, Os02g0152400; OsRbcS2, Os12g0274700; OsRbcS3, Os12g0291100; OsRbcS4, Os12g0292400; OsRbcS5, Os12g0291400; Selaginella moellendorffii 1, Sm147927; S. moellendorffii 2, Sm186173; S. moellendorffii 3, Sm017395; S. moellendorffii 4, Sm236158; grape 1, GSVIVP00017679001; grape 2, GSVIVP00031354001; and Populus trichocarpa, XP_002325043.
Expression of OsRbcS1
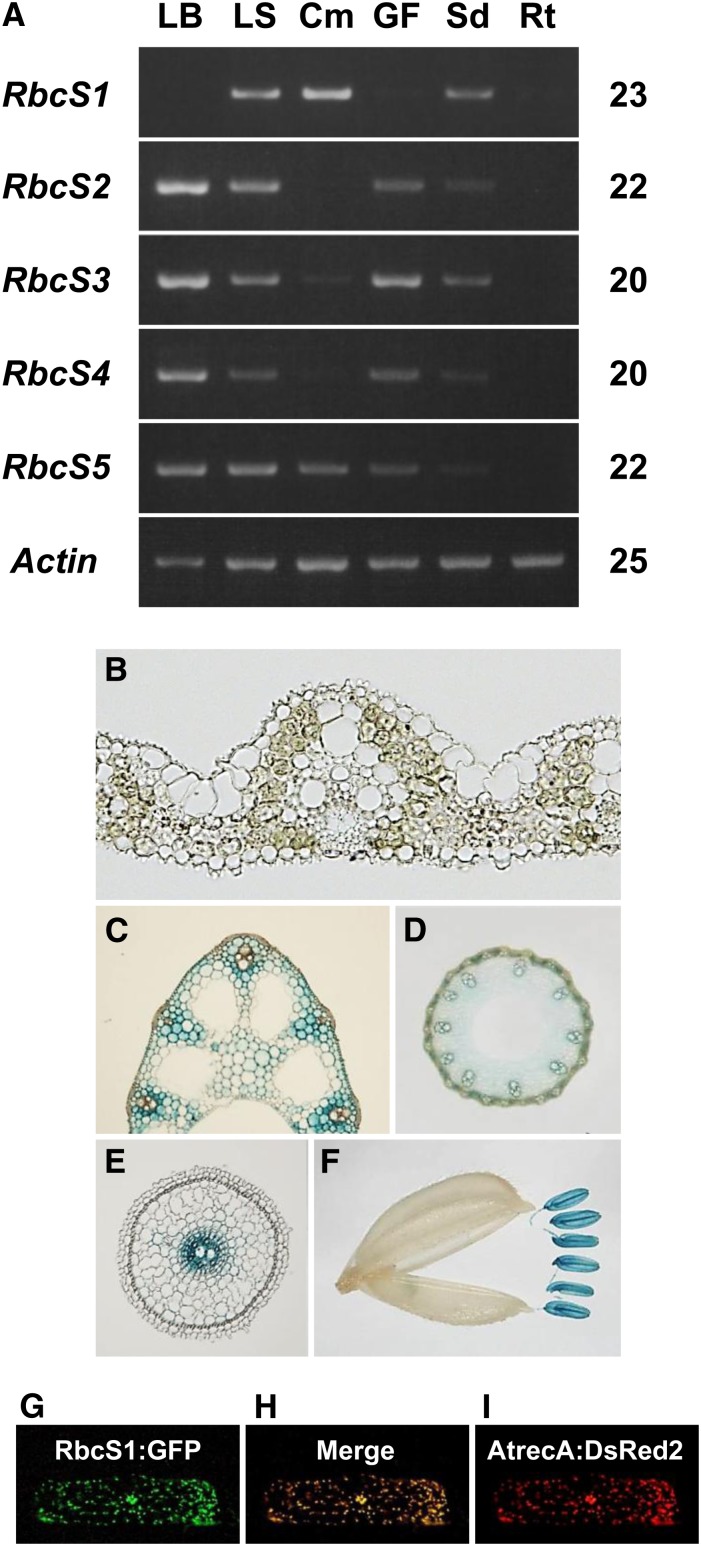
Reverse transcription (RT)-PCR analysis revealed that OsRbcS2 to OsRbcS5 showed a similar expression pattern of organ specificity, i.e. expressed in green tissues such as leaf blade, leaf sheath, and glumaceous flower but not in root (Fig. 2A). However, OsRbcS1 expression was not detectable in leaf blade, which is the major photosynthetic tissue in rice, but was detected in leaf sheath, culm, and developing seed. Thus, the organ specificity of the expression of OsRbcS1 was considered to be significantly different from those of other rice RbcSs. These observations are largely consistent with the results reported by Suzuki et al. (2009).
Figure 2.
Expression of OsRbcS1. A, Expression of rice RbcS gene family in various organs analyzed by RT-PCR. The numbers of PCR cycles are indicated on the right. The rice actin gene was also analyzed as an internal control. LB, Leaf blade; LS, leaf sheath; Cm, culm; GF, glumaceous flower; Sd, seed; Rt, root. B to F, Histochemical analysis of OsRbcS1:GUS expression. Cross sections of leaf blade (B), leaf sheath (C), culm (D), and root (E). Glumaceous flower (F). G to I, Subcellular localization of OsRbcS1. The chimeric construct, OsRbcS1:GFP (G), was expressed in onion epidermal cells. The construct of AtrecA:DsRed2 was used as a positive control for plastid localization (I). In the merged image, the colocalized signals of GFP and DsRed2 appear yellow (H).
Promoter GUS analysis was carried out to analyze the cell- and tissue-specific expression of OsRbcS1 (Fig. 2, B–F). OsRbcS3 was also analyzed, as representative rice RbcS reported to be highly expressed in photosynthetic tissues (Suzuki et al., 2009). The OsRbcS3 promoter GUS fusion was significantly expressed in mesophyll cells of leaf blade but not expressed in the basal, pale-green part of leaf sheath (Supplemental Fig. S4). By contrast, the expression of the OsRbcS1 promoter GUS fusion was not observed in leaf blade at all but could be found in cells around vascular bundles of pale-green leaf sheath (Fig. 2, B and C). The expression was also observed in upper green parts of leaf sheath. In addition, the expression of OsRbcS1 was detectable in culm, anther, filament, and central cylinder of young root (Fig. 2, D–F). All of these expressions are essentially confined to nonphotosynthetic cells, implying that OsRbcS1 might not be involved in photosynthesis.
The putative transit peptide of OsRbcS1 shows a relatively low degree of identity with other rice RbcSs and lacks a common amino acid motif ([P/G]Xn[R/K]Xn[S/T]Xn[S*/T*], where the asterisk represents the phosphorylation site and n = zero to three amino acids spacer) conserved in some nuclear-encoded chloroplast proteins including RbcS (Supplemental Fig. S1; Waegemann and Soll, 1996). Thus, it is uncertain whether OsRbcS1 could actually be targeted to the chloroplast. To examine the subcellular localization of OsRbcS1, a fusion protein of the N-terminal putative transit peptide of OsRbcS1 and GFP (OsRbcS1:GFP) was expressed in epidermal cells of onion (Allium cepa; Fig. 2, G–I). As a positive control for plastid targeting, a fusion protein AtrecA:DsRed2 (for Arabidopsis chloroplast DNA repair protein:Discosoma spp. Red fluorescent protein2; Imaizumi-Anraku et al., 2005) was expressed together with OsRbcS1:GFP. The GFP fluorescent completely overlapped with DsRed2 fluorescent. These findings indicate that the OsRbcS1 protein should be localized to the plastid, and the phosphorylation motif missing in OsRbcS1 is not always necessary for RbcS targeting to the chloroplast.
Overexpression of OsRbcS1 in Photosynthetic Tissues of Rice
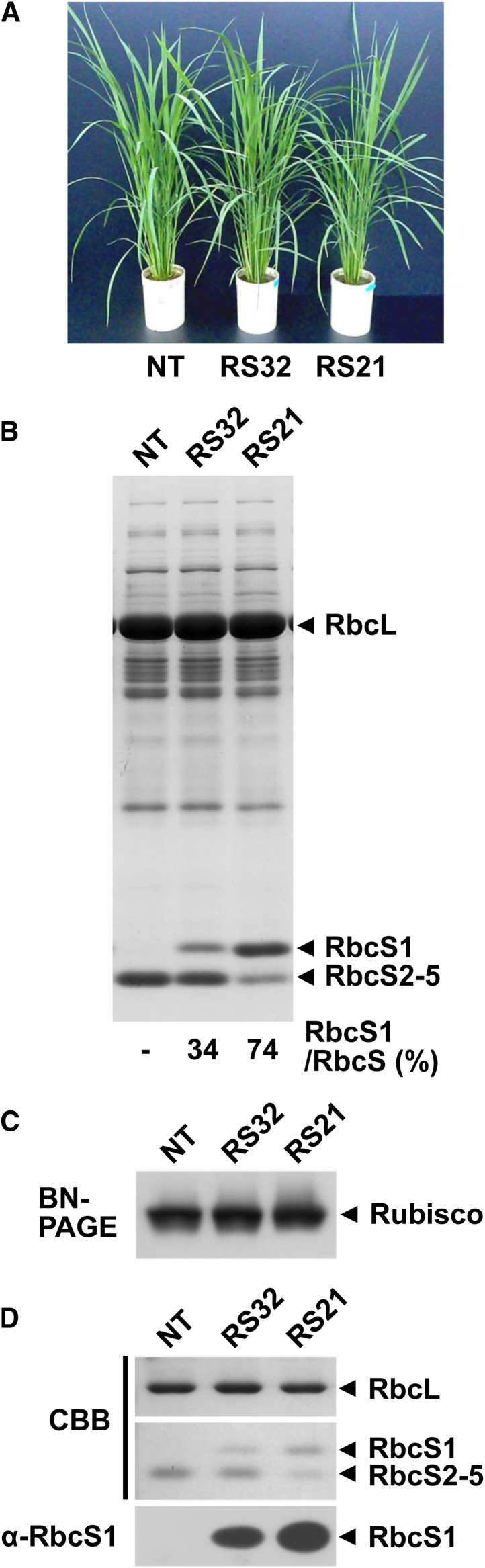
Transgenic rice overexpressing OsRbcS1 was generated by Agrobacterium tumefaciens-mediated transformation. The expression of the OsRbcS1 transgene was driven by the chlorophyll a/b-binding protein promoter, which directs green tissue-specific expression (Tada et al., 1991). Because the molecular mass of OsRbcS1 protein is greater than other rice RbcSs, these proteins could be separated by SDS-PAGE. Exploiting this difference in molecular mass, the expression level of OsRbcS1 in leaf blade of transgenic rice was estimated by SDS-PAGE with Coomassie Blue staining. The band of OsRbcS1 was present in most primary transgenic lines. Among these progenies, two homozygous transgenic lines, designated RS32 and RS21, showing Mendelian inheritance and thought to contain the transgene(s) at a single locus, were selected for subsequent experiments (Fig. 3A). The expression level of OsRbcS1 as a proportion of total rice RbcS in lines RS32 and RS21 was 34% and 74%, respectively (Fig. 3B). The expression level of OsRbcS2 to OsRbcS5 was reduced in proportion to the increase in OsRbcS1 in transgenic rice lines.
Figure 3.
Overexpression of OsRbcS1 in green tissues of transgenic rice. A, Transgenic rice plants overexpressing OsRbcS1 grown in soil at 37 d after transplanting. B, Expression levels of OsRbcS1 analyzed by SDS-PAGE. Total soluble proteins (6.4 µg protein) in leaf blade were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. Intensities of RbcS bands were quantified using the National Institutes of Health image program. The mean ratios of OsRbcS1 to total rice RbcS of five biological replicates (means ± sd) were 33.6 ± 0.77 and 74.1 ± 5.25 for RS32 and RS21, respectively. These values were significantly different from each other (P < 0.01). The total amount of RbcS in nontransgenic rice, RS32, and RS21 were 0.65, 0.68, and 0.74 μg, respectively. C, Analysis of Rubisco by blue native-PAGE. Total soluble proteins (2.0 µg protein) in leaf blade were separated by a 3% to 12% gradient gel and stained with Coomassie Brilliant Blue. D, SDS-PAGE and immunoblotting of Rubisco separated by blue native-PAGE. Rubisco bands from blue native-PAGE were excised from the gel and further separated by SDS-PAGE. Rubisco proteins were detected by Coomassie Brilliant Blue staining and immunoblotting with an antiserum raised against OsRbcS1. BN, Blue native; CBB, Coomassie Brilliant Blue; NT, nontransgenic rice. [See online article for color version of this figure.]
The subunit composition of Rubisco in leaf blade of transgenic rice was analyzed by blue native-PAGE and subsequent SDS-PAGE and immunoblotting (Fig. 3, C and D). Blue native-PAGE showed that the apparent molecular mass of Rubisco in transgenic lines was almost identical to that of nontransgenic rice (Fig. 3C). After blue native-PAGE, the Rubisco bands were excised from the gel and further separated by SDS-PAGE. Detection of Rubisco bands by Coomassie Blue staining and immunoblotting demonstrated that OsRbcS1 band was only present in transgenic lines (Fig. 3D). These results suggested that OsRbcS1 was correctly incorporated into L8S8 Rubisco in leaves of transgenic rice where Rubisco was present as a chimera of OsRbcS1 and other rice RbcS.
OsRbcS1 expression was detected in some tissues such as leaf sheath and culm of nontransgenic rice (Fig. 2). However, it was still uncertain that OsRbcS1 protein functioned as a small subunit in Rubisco of nontransgenic rice. To resolve this question, subunit composition of Rubisco expressed in leaf sheath of nontransgenic rice was also studied (Fig. 4). Although the expression of Rubisco containing OsRbcS1 would be dominant in the basal, pale-green part of leaf sheath, its expression level was presumed to be considerably low compared with Rubisco in green tissues. Therefore, the greener part of leaf sheath expressing OsRbcS1 as well as other rice RbcSs were used for the analysis to facilitate the excision of the Rubisco band from blue native-PAGE gel. The band of OsRbcS2 to OsRbcS5 was detected both in leaf blade and leaf sheath by immunoblotting, even though antibodies raised against OsRbcS1 were used for detection, suggesting that Rubisco in these tissues contained considerable amount of OsRbcS2 to OsRbcS5. By contrast, OsRbcS1 band could only be detected from Rubisco expressed in leaf sheath but not in leaf blade. These results indicate that OsRbcS1 is actually incorporated into Rubisco and functions as Rubisco small subunit in some tissues of rice.
Figure 4.
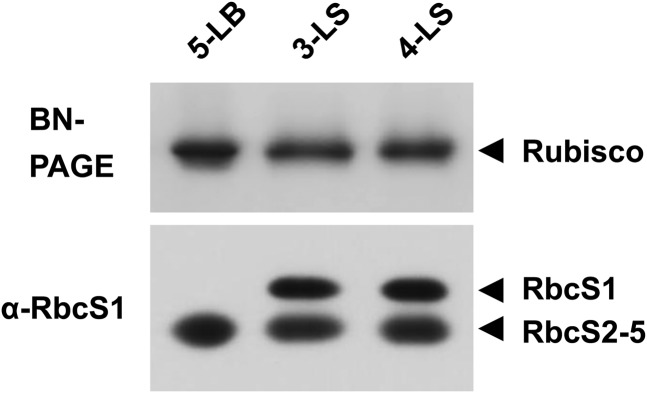
Subunit composition of Rubisco in nontransgenic rice. Total soluble proteins of leaf blade and leaf sheath (2.0 µg for leaf blade and 4.0 µg for leaf sheath) at 5.5 leaf stage were separated on a 3% to 12% gradient gel by blue native-PAGE and stained with Coomassie Brilliant Blue (top). Rubisco bands from blue native-PAGE were excised from the gel and further separated by SDS-PAGE. RbcS proteins were detected by immunoblotting with an antiserum raised against OsRbcS1 (bottom). BN, Blue native; 5-LB, fifth leaf blade; 3-LS, third leaf sheath; 4-LS, fourth leaf sheath.
The effects of overexpression of OsRbcS1 in green tissues on growth of transgenic rice plants were also examined. No remarkable differences in plant length, leaf number, and tiller number were observed between transgenic and nontransgenic rice plants, with the exception of a slight decrease in tiller number in a high expression line RS21, compared with nontransgenic rice (Supplemental Fig. S5). Also, panicle and shoot dry weight were significantly decreased only in RS21 compared with nontransgenic rice (Supplemental Fig. S6), possibly due to the decreased tiller number.
Catalytic Properties of Rubisco Containing OsRbcS1
Because the deduced amino acid sequence of OsRbcS1 was significantly different from those of other rice RbcSs (Supplemental Figs. S1 and S2), it was expected that the Rubisco containing OsRbcS1 may have altered catalytic properties. The kcats of Rubisco of transgenic lines RS32 and RS21 were 1.16- and 1.46-fold higher than that of nontransgenic rice (Fig. 5). Km for CO2 of Rubisco (Kc) in transgenic lines also tended to be higher than that of nontransgenic rice. Increase in kcat and Kc of Rubisco observed in transgenic lines were largely correlated with the expression level of OsRbcS1. However, the extent of increase in Kc, particularly of RS21, a high expression line, was much larger than that in kcat, showing more than 3-fold higher Kc compared with nontransgenic rice. The CO2/O2 specificity of Rubisco (Sc/o) of transgenic line RS21 was lower than that of nontransgenic rice. These results indicate that the overexpression and the incorporation of OsRbcS1 into Rubisco shift its catalytic properties to a high-activity-type Rubisco with a higher kcat and lower affinity for CO2, as found in C4 plants and cold-resistant plants (Sage, 2002; Ishikawa et al., 2009).
Figure 5.
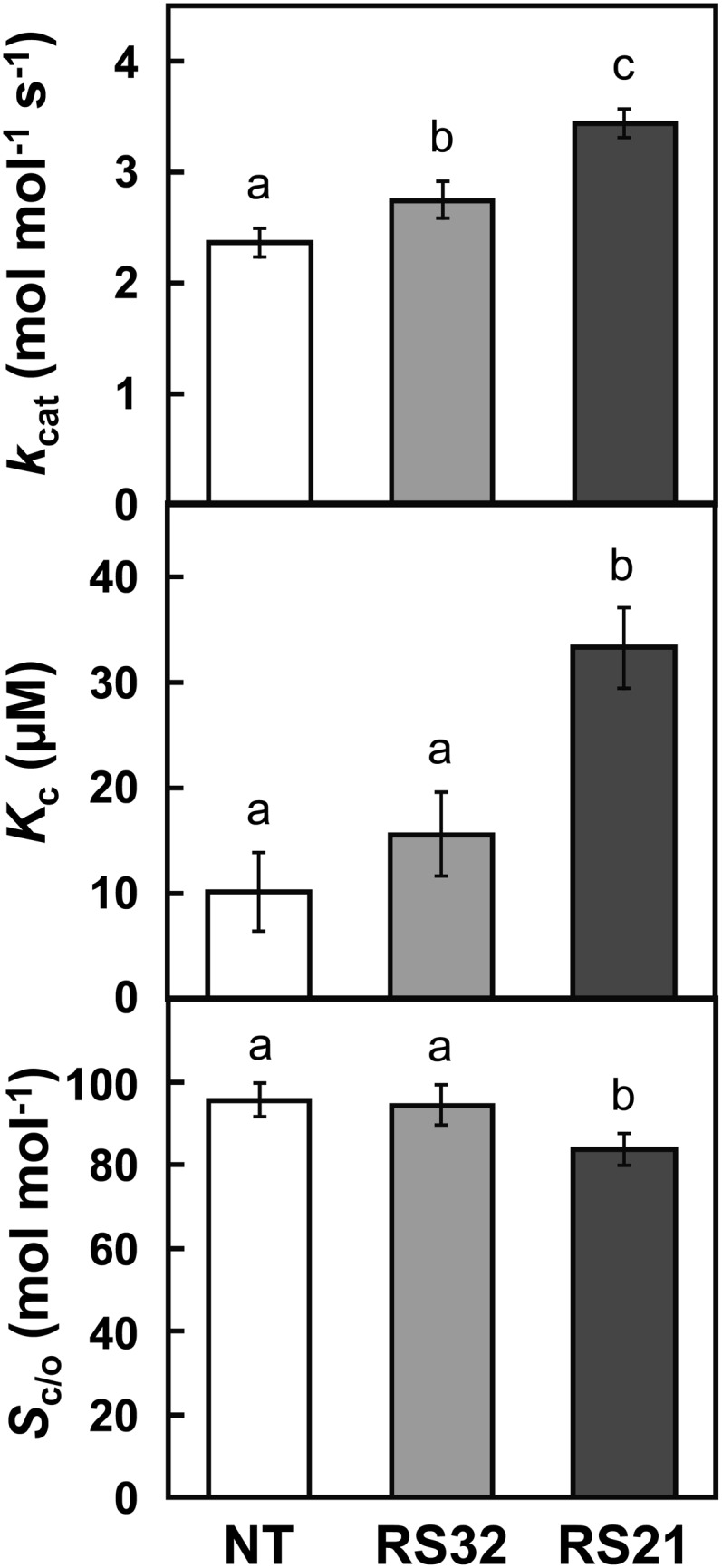
Kinetic properties of Rubisco. Sc/o was determined by the measurement of gas exchange (see “Materials and Methods” for details). Data represent means ± sd of five biological replicates. Different letters above the bars indicate significant difference (P < 0.05) between lines determined by Tukey’s test. NT, Nontransgenic rice.
Photosynthesis and Leaf Constituents of Transgenic Rice
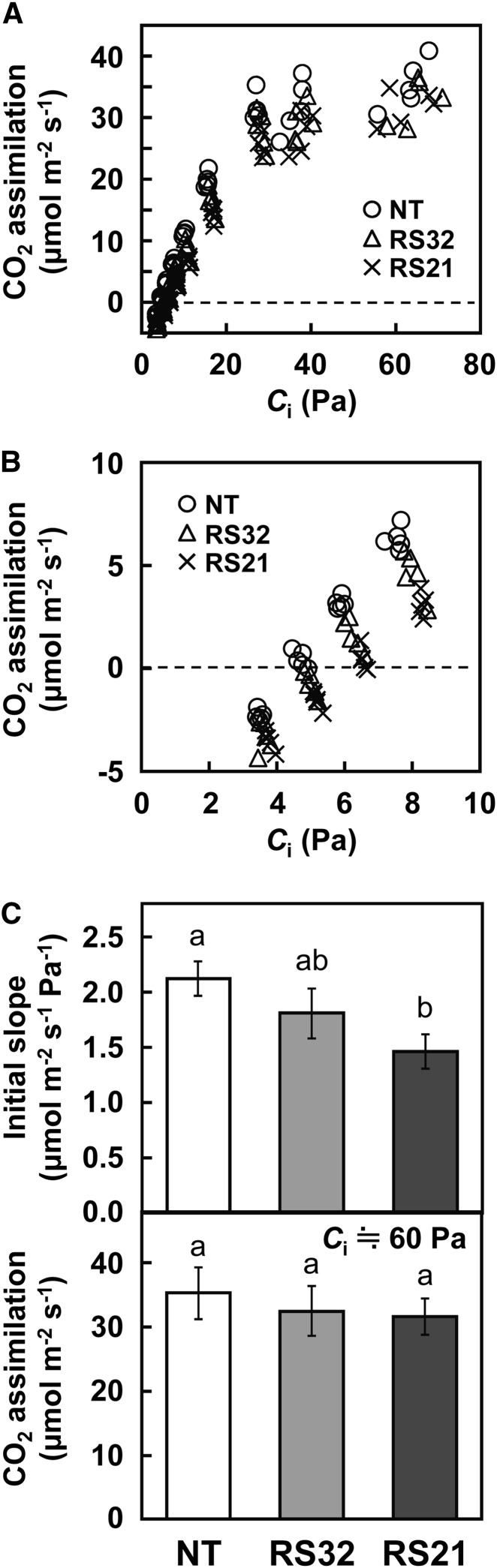
The CO2 assimilation rate as a function of intercellular partial pressure of CO2 inside leaf (Ci) was measured in two transgenic rice lines (Fig. 6A). The CO2 assimilation rates in both transgenic lines were similar at ambient to higher CO2 partial pressures, whereas they tended to be lower with decreasing Ci compared with that of nontransgenic rice. The initial slopes of the CO2 assimilation rate/Ci curve, an indicator of Rubisco-limited photosynthetic capacity, were decreased in transgenic lines compared with nontransgenic rice (Fig. 6, B and C). By contrast, the CO2 assimilation rate under high Ci (approximately 60 Pa), in which photosynthetic rate would be largely limited by RuBP regeneration capacity, was unchanged (Fig. 6C). Using the data of CO2 assimilation rate and kinetic properties of Rubisco, the maximum rate of RuBP carboxylation (Vcmax) and the maximum rate of photosynthetic electron transport (Jmax) were estimated (Supplemental Fig. S7). Although the initial slopes of the CO2 assimilation rate/Ci curve were decreased, Vcmax in transgenic lines was substantially higher than that in nontransgenic rice. These results are likely due to significant changes in the kinetic properties of Rubisco to high-activity type in transgenic lines (Fig. 5). By contrast, consistent with the results of the CO2 assimilation rate at higher CO2 partial pressure, Jmax did not differ among genotypes. Because the Vcmax was substantially increased, the Jmax/Vcmax ratio was significantly decreased in transgenic lines, suggesting that the capacity of photosynthetic electron transport was insufficient to support the capacity of RuBP carboxylation enhanced by the introduction of OsRbcS1 in transgenic rice under higher CO2 conditions.
Figure 6.
Photosynthetic rate of transgenic rice overexpressing OsRbcS1. A, Dependence on Ci of photosynthetic CO2 assimilation rate. The CO2 assimilation rate of the uppermost fully expanded leaves at vegetative growth stage was measured at a PPFD of 1,500 µmol m–2 s–1, leaf temperature of 28°C, and CO2 partial pressures ranging from 4.5 to 88 Pa. B, Magnified figure of Figure 6A at low Ci. C, Initial slope of CO2 assimilation rate as a function of Ci and CO2 assimilation rate at Ci of approximately 60 Pa. The values were calculated by the data of Figure 6B. Data represent means ± sd of five biological replicates. Different letters above the bars indicate significant difference (P < 0.05) between lines determined by Tukey’s test. NT, Nontransgenic rice.
The Rubisco content was increased by 13% to 21% in transgenic lines compared with nontransgenic rice, and its increase was dependent on the expression levels of OsRbcS1 (Fig. 7). With this increase in Rubisco content, the soluble protein content was marginally increased in transgenic lines. By contrast, the chlorophyll content was decreased in the high-expression line RS21. Although the contents of major leaf constituents such as Rubisco and chlorophyll varied, there were no significant differences in the leaf total nitrogen content between transgenic and nontransgenic rice.
Figure 7.
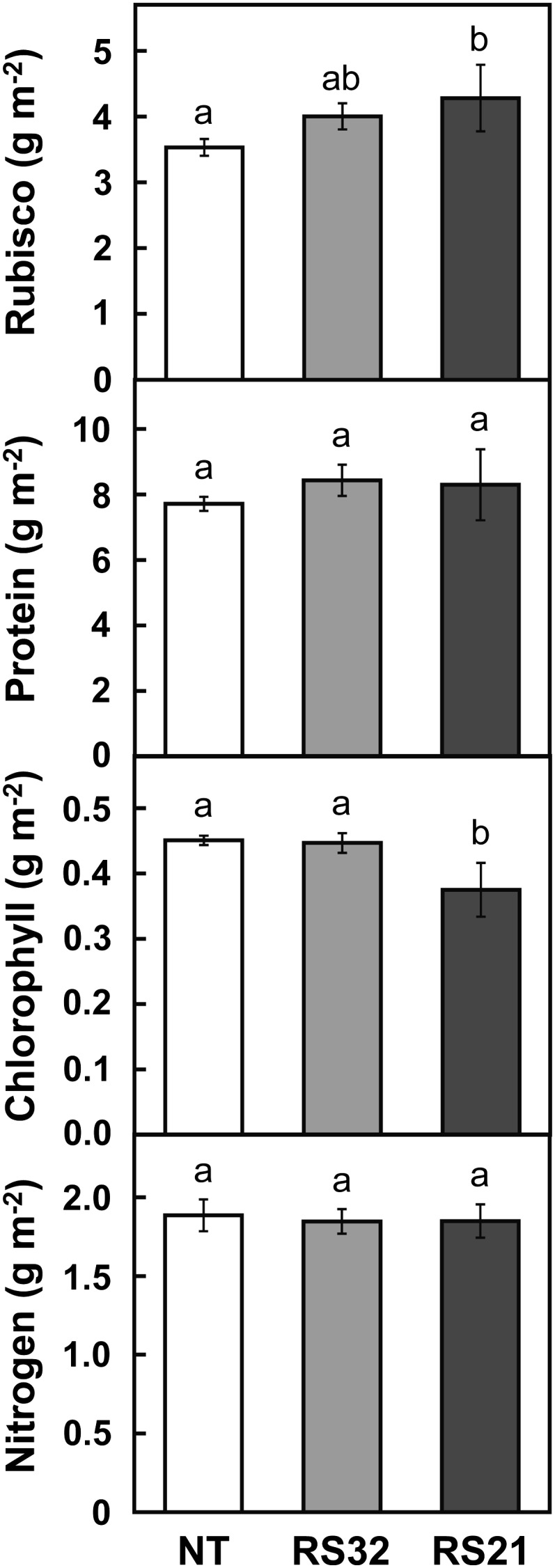
Rubisco, soluble protein, chlorophyll, and nitrogen contents in leaves of transgenic rice overexpressing OsRbcS1. The uppermost fully expanded leaves were collected after the measurement of photosynthetic rate. Data represent means ± sd of five biological replicates. Different letters above the bars indicate significant difference (P < 0.05) between lines determined by Tukey’s test. NT, Nontransgenic rice.
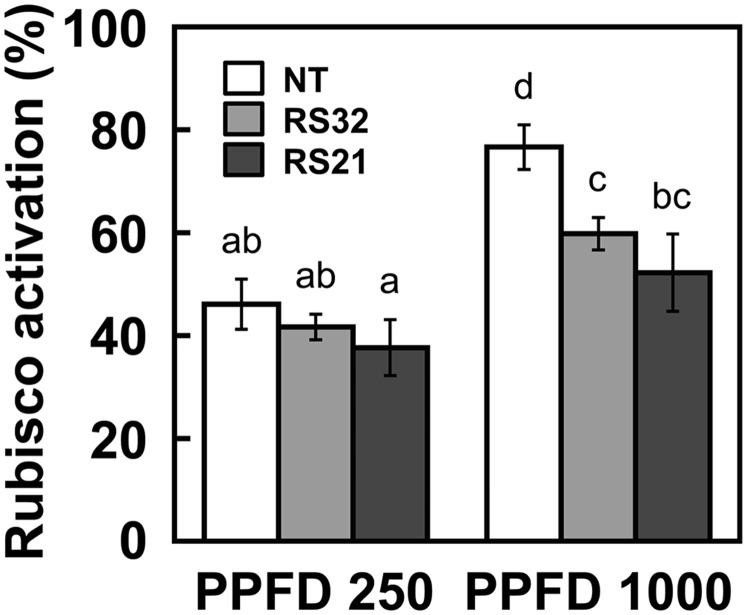
The activation state of Rubisco in leaves was decreased in transgenic lines (Fig. 8). The decreases in Rubisco activation were more marked under high light intensity than under low light intensity and were largely correlated with the expression levels of OsRbcS1. As shown in Figure 3B, Rubisco in line RS21 was composed of more than 70% OsRbcS1. Considering the expression level of OsRbcS1 in leaves of transgenic lines, it is apparent that Rubisco containing OsRbcS1 can be activated by Rubisco activase expressed in photosynthetic tissues. Because the Jmax/Vcmax ratio was significantly decreased in the transgenic lines (Supplemental Fig. S7), the observed lower Rubisco activation might be caused by the imbalance between the capacities of RuBP carboxylation and photosynthetic electron transport, as previously observed in transgenic rice overexpressing sorghum RbcS (Ishikawa et al., 2011).
Figure 8.
The activation state of Rubisco in leaves of transgenic rice overexpressing OsRbcS1 under different light intensities. Rice leaves were collected under the irradiation of PPFD of 250 or 1,000 µmol m–2 s–1, leaf temperature of 28°C, and CO2 partial pressure of 38 Pa. The activation state of Rubisco was analyzed by measuring Rubisco activity. Data represent means ± sd of five biological replicates. Different letters above the bars indicate significant difference (P < 0.05) between lines determined by Tukey’s test. NT, Nontransgenic rice.
DISCUSSION
In this study, we focused on OsRbcS1, the only member of five RbcS in rice that showed remarkably low deduced amino acid sequence identity with the other RbcS members of the gene family. This unique amino acid sequence raises the question whether OsRbcS1 could be correctly assembled with rice RbcL to form a functional Rubisco holoenzyme. We found that OsRbcS1 was targeted to the chloroplast and incorporated into L8S8 Rubisco in the leaf blade of transgenic rice overexpressing OsRbcS1 (Figs. 2 and 3). Likewise, OsRbcS1 was present in Rubisco of nontransgenic rice in leaf sheath where OsRbcS1 expression level was relatively high (Fig. 4). These results clearly demonstrate that OsRbcS1 can function as the small subunit of Rubisco in rice.
In a previous study, sorghum RbcS expressed in rice was successfully assembled with rice RbcL to form functional Rubisco (Ishikawa et al., 2011). Also, functional incorporations of sunflower (Helianthus annuus) RbcL or Flaveria spp. RbcL with tobacco RbcS have been reported in transgenic tobacco (Sharwood et al., 2008; Whitney et al., 2011). Moreover, it has been reported that even more genetically distant combinations such as RbcL from cyanobacteria and algae could assemble with RbcS from vascular plants to form functional hybrid Rubisco (van der Vies et al., 1986; Wang et al., 2001; Genkov et al., 2010). These observations imply that a wide range of foreign RbcS can be processed during Rubisco assembly in nature. Additionally, most of the highly conserved amino acid residues among RbcS are present in OsRbcS1 (Supplemental Fig. S2). These may enable markedly different RbcS proteins, namely, OsRbcS1 and other rice RbcSs, to function in a plant.
Because the catalytic sites of Rubisco reside in RbcL, there is no doubt that RbcL plays a crucial role in the catalysis of the reaction. Consistent with this, the amino acid sequences of RbcL are highly homologous among photosynthetic organisms containing L8S8 Rubisco, whereas RbcS shows much more diversity than RbcL (Spreitzer, 2003). This implies that RbcS might be related to the natural variation of the catalytic properties of Rubisco observed among photosynthetic organisms. Recently, Ishikawa et al. (2011) showed that the introduction of RbcS of a high kcat Rubisco from sorghum significantly increased the kcat and Kc of rice Rubisco. Similarly, substantial effects on the catalytic properties of Rubisco were also observed when OsRbcS1 was incorporated into rice Rubisco in this study (Fig. 5). These results strongly support the hypothesis that RbcS can make a significant contribution to determining the catalytic performance of Rubisco. In the amino acid sequence of RbcS, the βA-βB loop is highly variable among species and has been suggested to be important for Rubisco catalytic function (Spreitzer, 2003). In this region, nine amino acid residues of OsRbcS1 differ from those of other rice RbcSs (Supplemental Fig. S2). Among these amino acid substitutions, two of the nine are common to sorghum RbcS. These are Val-49 and His-57 in OsRbcS2 to OsRbcS5 and Glu and Ser in OsRbcS1 and sorghum RbcS, respectively. Although functional significance has not yet been demonstrated, these amino acid substitutions may affect the catalytic properties of Rubisco observed in our transgenic lines.
Previous studies have reported that the overexpression of native or sorghum C4 RbcS in transgenic rice did not affect photosynthetic rate under any given CO2 partial pressures (Suzuki et al., 2007; Ishikawa et al., 2011). Consistent with those observations, the CO2 assimilation rate under ambient to higher CO2 conditions did not differ between OsRbcS1 overexpression lines and nontransgenic rice (Fig. 6). However, in this study, the overexpression of OsRbcS1 reduced the photosynthetic rate under low CO2 conditions compared with nontransgenic rice (Fig. 6, B and C). These observations suggest that Rubisco catalysis under low CO2 conditions is compromised in transgenic rice. Rubisco from these transgenic rice lines showed a higher Kc and slightly lower Sc/o than those in nontransgenic rice (Fig. 5). These catalytic properties would be expected to be disadvantageous for CO2 fixation under low CO2 conditions. Sorghum RbcS had similar effects on the kinetic properties and the expression level of Rubisco with OsRbcS1, whereas the effect of OsRbcS1 on Kc was larger than that of sorghum RbcS: Kc in transgenic rice that expresses sorghum RbcS was between 1.25- and 1.39-fold higher, while in rice expressing OsRbcS1, Kc was 1.54- to 3.27-fold higher than in nontransgenic rice (Fig. 5; Ishikawa et al., 2011). This large effect on Kc could be responsible for the decrease in photosynthetic rate seen here under low CO2 conditions in OsRbcS1 overexpression lines.
It has been reported in many plant species that the expression of individual members of the RbcS gene family is differently regulated depending on organ, developmental stage, and environmental conditions such as light and temperature (Dean et al., 1989; Silverthorne and Tobin, 1990; Wanner and Gruissem, 1991; Dedonder et al., 1993; Peters and Silverthorne, 1995; Yoon et al., 2001). Given the high homology between the amino acid sequences of these gene family members, it is difficult to infer functional significance to this observation. In addition, the expression of RbcS is basically confined to photosynthetic tissues such as leaf. It has been assumed that plants contain multiple RbcS genes to achieve high-level expression and to modulate their expression level in response to various signals. In rice, the deduced amino acid sequence and the organ specificity of the expression of OsRbcS1 were substantially different from those of the other members of RbcS gene family (Fig. 2; Supplemental Figs. S1 and S2). Moreover, the incorporation of OsRbcS1 into Rubisco significantly altered the catalytic properties of the enzyme (Fig. 5). These results imply that rice utilizes functionally distinct RbcS isoforms to alter the catalytic properties of Rubisco depending on the tissues and the conditions in which they function. As described in “Results,” OsRbcS1-like genes are also present in plants other than rice. Detailed expression analyses of these genes will be of interest in understanding their functional significance.
OsRbcS1 expresses and functions as the small subunit of Rubisco in leaf sheath of nontransgenic rice but not in leaf blade (Figs. 2 and 4). This observation raises the possibility that Rubisco containing OsRbcS1 did not evolve to function in normal C3 photosynthetic CO2 fixation. Rubisco has been reported to function in some metabolic pathways other than the Calvin cycle and in the refixation of CO2 in particular tissues; for example, C4-like photosynthesis can be operative in the cells around vascular bundles in stems and petioles of some C3 species (Hibberd and Quick, 2002), and in developing seeds and siliques of some oil-producing plant species, Rubisco is considered to contribute significantly to the refixation of CO2 released during oil biosynthesis (Ruuska et al., 2004). It was also proposed that Rubisco is involved in fatty acid biosynthesis to improve carbon use efficiency without operation of a full Calvin cycle in Brassica napus (Schwender et al., 2004). In these tissues, CO2 concentrations would be higher than that in mesophyll cells when photosynthetic CO2 fixation is operative. The kinetic properties of Rubisco containing OsRbcS1 could be more suitable for catalysis in these high CO2 environments than those of Rubisco with other rice RbcSs.
In this study, we report the first demonstration of an RbcS encoded by RbcS gene family significantly altering catalytic properties of Rubisco. This unusual RbcS, designated OsRbcS1, increases kcat and Kc to become a high-activity-type Rubisco similar to that found in photosynthetic organisms such as algae and C4 plants whose Rubisco is adapted to function in high CO2 environments. A catalytic “tradeoff” is often observed between kcat and Kc (Ishikawa et al., 2009) or kcat and Sc/o (Tcherkez et al., 2006). Hence, increasing kcat is likely to lead to a decrease in the affinity and specificity for CO2. However, considering the recent rapid rise in atmospheric CO2 concentration, a high-activity-type Rubisco is expected to provide potential improvement of photosynthetic performance for C3 plants in the future. In a previous study, we successfully increased the kcat of rice Rubisco by the introduction of sorghum RbcS (Ishikawa et al., 2011). To enhance the kcat of Rubisco, we propose that OsRbcS1 could be a much better candidate than sorghum RbcS, because native RbcS would be expected to be more amenable to biosynthesis, turnover, and activation of Rubisco holoenzyme in transgenic plants. However, the expected enhancement of photosynthetic rate was not achieved here by the overexpression of OsRbcS1 alone (Fig. 6). Makino et al. (1997) estimated that Rubisco content could be in excess at elevated CO2, and it could be reduced to improve nitrogen use efficiency without compromising photosynthetic rate. A reduction of Rubisco content would allow a greater nitrogen investment in the other photosynthetic components. Therefore, engineering both the catalytic properties and the optimization of content of Rubisco will be required to improve photosynthesis and eventually agricultural productivity in C3 plants. OsRbcS1 may not currently be an important gene for rice photosynthesis but may become more important for rice of the future.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa ‘Nipponbare’) plants were grown in soil under natural sunlight conditions in a temperature-controlled greenhouse at day and night temperatures of 30°C and 23°C, respectively, and relative humidity of 70%, as described previously (Fukayama et al., 2012). All samples for molecular and biochemical analyses were collected at 11 am to 1 pm on sunny days, immediately frozen in liquid nitrogen, and stored at –80°C until required.
Sequence Alignment and Phylogenetic Analysis
The information of amino acid sequences of RbcS were obtained from the RAP-DB (http://rapdb.dna.affrc.go.jp), National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), and the Surveyed Conserved Motif Alignment Diagram and the Associating Dendrogram (SALAD) database (http://salad.dna.affrc.go.jp/salad). The sequences were aligned using ClustalX software (http://www.clustal.org/clustal2) and imported into NJplot software (http://pbil.univ-lyon1.fr/software/njplot.html) to construct a phylogenetic tree. The tree was constructed using the neighbor-joining method.
Plasmid Construction and Transformation of Rice
All primers used for plasmid construction are listed in Supplemental Table S1. To construct promoter:GUS chimeric genes, 5′-flanking regions of OsRbcS1 (from –1,044 to +218, numbered from the translation initiation site) and OsRbcS3 (from –1,000 to +68) were amplified from rice genomic DNA by PCR and inserted upstream of GUS in the binary vector pBI-Hm (a generous gift from Makoto Matsuoka [Nagoya University]). To construct the OsRbcS1:GFP fusion protein, the nucleotide sequence encoding the putative transit peptide of the OsRbcS1 (amino acid residues 1–50; Supplemental Fig. S1) was amplified from a first-strand complementary DNA (cDNA) of leaf sheath by PCR and cloned into a GFP expression vector (CaMV35S-sGFP[S65T]-nos3′; a generous gift from Yasuo Niwa [University of Shizuoka]; Chiu et al., 1996). For overexpression of OsRbcS1, a full-length cDNA of OsRbcS1 was amplified from the first-strand cDNA by PCR and cloned into the binary vector pIG121Hm (a generous gift from Kenzo Nakamura [Chubu University]) containing the rice chlorophyll a/b-binding protein promoter. Rice transformation was performed via Agrobacterium tumefaciens-mediated gene transfer. Antibiotic-resistant transgenic rice plants (T1 generation) were regenerated and grown in soil. Among these progeny (T2 generation), two homozygous transgenic lines, designated RS32 and RS21, showing a segregation ratio of around 1:3 were selected and used for subsequent experiments.
RT-PCR
Total RNA was extracted from rice tissues using the RNeasy Plant Mini Kit (Qiagen). First-strand cDNA was synthesized using the PrimeScriptII First-Strand cDNA Synthesis Kit (Takara). Semiquantitative RT-PCR was performed using Quick Taq HS DyeMix (Toyobo) with 5% (v/v) dimethyl sulfoxide and the primers listed in Supplemental Table S1. The DNA polymerase was first activated at 94°C for 2 min, and PCR was carried out for 20 to 25 cycles of 30 s at 94°C, 30 s at 56°C to 62°C, and 30 to 50 s at 68°C, followed by a final extension step for 7 min at 68°C.
Promoter GUS and GFP Analysis
Histochemical analysis of GUS activity was performed as described in Matsuoka and Numazawa (1991). Plant tissues were incubated with GUS staining solution (50 mm sodium phosphate, 0.5 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 0.5 mm K3Fe(CN)6, 0.5 mm K4Fe(CN)6, and 0.05% [v/v] Triton X-100, pH 7.0) for 3 to 8 h at 37°C. An ethanol wash was performed to stop the GUS reaction and remove chlorophyll from the tissues.
The GFP expression vector was introduced into the epidermis of onion (Allium cepa) by particle bombardment using 1-µm gold particles and a gene delivery system (PDS-1000, Bio-Rad). As a positive control for plastid targeting, a vector containing a chimeric construct, AtrecA:DsRed2 (Imaizumi-Anraku et al., 2005), was introduced together with the GFP expression vector. After incubation in darkness for 48 h at room temperature, the epidermis was peeled from the bombarded segment, and GFP and DsRed2 activity were observed with a fluorescent microscope (BX50, Olympus) through U-MNIBA and U-MWIG cubes (Olympus), respectively.
Electrophoresis and Immunoblotting
Total soluble proteins were extracted from rice tissues in extraction buffer containing 50 mm HEPES-KOH, 10 mm MgCl2, 1 mm EDTA, 5 mm dithiothreitol (DTT), 0.01 mm leupeptin, 1 mm phenylmethylsulfonylfluoride, 10% (w/v) glycerol, and 5% (w/v) polyvinylpolypyrrolidone, pH 7.4, with a small amount of quartz sand. For blue native-PAGE, 20 mm ascorbate, 4 mm amino-n-capronic acid, and 0.8 mm benzamidine were added to the extraction buffer. The homogenates were centrifuged at 15,000 rpm for 5 min at 4°C. SDS-PAGE (14%) was performed as described previously (Masumoto et al., 2010). Blue native-PAGE was carried out using a precast gradient gel (NativePAGE Novex 3%–12% Bis-Tris Gel, Life Technologies) according to the manufacturer’s instructions. Rubisco bands of blue native-PAGE were excised from the gel and incubated in equilibrium buffer containing 62.5 mm Tris-HCl, 2% (w/v) DTT, 1% (w/v) SDS, and 6 m urea (pH 6.8). Proteins in the gels were further separated by SDS-PAGE. The gels were stained with Coomassie Brilliant Blue R-250 or subjected to immunoblotting using an antiserum raised against OsRbcS1. Immunoreacted bands were visualized with the ECL Advance Western Blotting Detection Kit (GE Healthcare) and exposed to x-ray film.
Rubisco Activity
For measurement of total activity of Rubisco, leaves were homogenized in extraction buffer containing 100 mm bicine-NaOH, 5 mm MgCl2, 1 mm EDTA, 2 mm NaH2PO4, 5 mm DTT, 4 mm amino-n-capronic acid, 0.8 mm benzamidine, 0.4% (w/v) bovine serum albumin, and 1% (w/v) polyvinylpolypyrrolidone, pH 8.0, using a chilled mortar and pestle. The homogenates were centrifuged at 12,000 rpm for 2 min at 4°C. The supernatants were used for determination of Rubisco activity and catalytic sites. Rubisco in the supernatant was activated by the incubation with 15 mm MgCl2 and 5 mm NaHCO3 for 15 min on ice. Rubisco activity was determined using [14C]NaHCO3 by assaying the incorporation of 14C into acid-stable product. The Rubisco was added to the reaction buffer (100 mm bicine-NaOH, 20 mm MgCl2, 1 mm EDTA, and 5 mm DTT, pH 8.2) containing 0.5 mm RuBP, 4.0 Wilbur-Anderson units mL–1 carbonic anhydrase, and 1 to 20 mm NaH14CO3. After 1 min, formic acid was added to stop the reaction. The kcat and Kc of Rubisco were determined and calculated by the Hanes-Woolf plot as described in Ishikawa et al. (2011). For measurement of initial activity of Rubisco, leaves were ground to a fine powder in liquid nitrogen using a mortar and pestle. The leaf powder was suspended in extraction buffer (50 mm bicine-NaOH, 100 µm EDTA, and 5 mm DTT, pH 7.8). After brief centrifugation, the supernatant was used for determination of Rubisco activity and catalytic sites. All reactions were performed at 28°C. The activation state of Rubisco was calculated as the ratio of initial activity to total activity.
Photosynthetic Rate
CO2 gas exchange rates of the uppermost fully expanded leaves of the main culm were measured with an open gas-exchange system (LI-6400, LI-COR). Measurements were performed at a photosynthetic photon flux density (PPFD) of 1,500 mol m–2 s–1, a leaf temperature of 28°C, and ambient CO2 partial pressures ranging from 4.5 to 88 Pa. Sc/o was determined by measurement of CO2 compensation points under different O2 partial pressures ranging from 2% to 20% (Laisk and Loreto, 1996).
Rubisco, Soluble Protein, Chlorophyll, and Nitrogen
Leaves were homogenized in extraction buffer containing 50 mm HEPES-KOH, 5 mm MgCl2, 1 mm EDTA, 5 mm DTT, 4 mm amino-n-capronic acid, 0.8 mm benzamidine, 5% (w/v) glycerol, 0.05% (v/v) Triton X-100, and 0.1% (w/v) polyvinylpolypyrrolidone, pH 7.5, using a chilled mortar and pestle. The homogenate was centrifuged at 15,000 rpm for 5 min at 4°C, and the supernatant was used for determination of Rubisco and total soluble protein. Rubisco was determined by assuming the stoichiometric binding of [14C]carboxyarabinitol bisphosphate as described in Ishikawa et al. (2011). Soluble protein was determined by the method of Bradford (1976) with bovine serum albumin as the standard. For determination of chlorophyll, an aliquot of the homogenate before centrifugation was taken from the mortar and extracted in 80% (v/v) acetone. Chlorophyll was determined spectrophotometrically as described in Porra et al. (1989). Total nitrogen was determined by indophenol colorimetric assay after the Kjeldahl digestion of leaf tissue as described previously (Fukayama et al., 2012).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of deduced amino acid sequences of the rice RbcS gene family.
Supplemental Figure S2. Alignment of amino acid sequences of mature protein of RbcS from rice, sorghum, spinach (Spinacia oleracea), and Chlamydomonas reinhardtii.
Supplemental Figure S3. DNA gel-blot analysis of RbcS among Poaceae.
Supplemental Figure S4. Histochemical analysis of OsRbcS3:GUS expression.
Supplemental Figure S5. Growth of transgenic rice plants overexpressing OsRbcS1.
Supplemental Figure S6. Panicle and shoot dry weight of transgenic rice plants overexpressing OsRbcS1.
Supplemental Figure S7. Jmax and Vcmax of transgenic rice overexpressing OsRbcS1.
Supplemental Table S1. The primers used in this study.
Acknowledgments
We thank Dr. Makoto Matsuoka (Nagoya University), Dr. Yasuo Niwa (University of Shizuoka), and Dr. Haruko Imaizumi-Anraku (National Institute of Agrobiological Sciences) for generous gifts of binary vector, GFP expression vector, and AtrecA:DsRed2 vector, respectively.
Glossary
- RuBP
ribulose-1,5-bisphosphate
- kcat
catalytic turnover rate
- RT
reverse transcription
- Kc
Km for CO2 of Rubisco
- Sc/o
CO2/O2 specificity of Rubisco
- Ci
partial pressure of CO2 inside leaf
- Vcmax
maximum rate of ribulose-1,5-bisphosphate carboxylation
- Jmax
maximum rate of photosynthetic electron transport
- DTT
dithiothreitol
- PPFD
photosynthetic photon flux density
- cDNA
complementary DNA
References
- Andersson I, Backlund A. (2008) Structure and function of Rubisco. Plant Physiol Biochem 46: 275–291 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Dean C, Pichersky E, Dunsmuir P. (1989) Structure, evolution, and regulation of RbcS genes in higher plants. Annu Rev Plant Physiol Plant Mol Biol 40: 415–439 [Google Scholar]
- Dedonder A, Rethy R, Fredericq H, Van Montagu M, Krebbers E. (1993) Arabidopsis rbcS genes are differentially regulated by light. Plant Physiol 101: 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JR. (1989) Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78: 9–19 [DOI] [PubMed] [Google Scholar]
- Evans JR, Austin RB. (1986) The specific activity of ribulose-1,5-bisphosphate carboxylase in relation to genotype in wheat. Planta 167: 344–350 [DOI] [PubMed] [Google Scholar]
- Fukayama H, Ueguchi C, Nishikawa K, Katoh N, Ishikawa C, Masumoto C, Hatanaka T, Misoo S. (2012) Overexpression of Rubisco activase decreases the photosynthetic CO2 assimilation rate by reducing Rubisco content in rice leaves. Plant Cell Physiol 53: 976–986 [DOI] [PubMed] [Google Scholar]
- Genkov T, Meyer M, Griffiths H, Spreitzer RJ. (2010) Functional hybrid Rubisco enzymes with plant small subunits and algal large subunits: engineered rbcS cDNA for expression in Chlamydomonas. J Biol Chem 285: 19833–19841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd JM, Quick WP. (2002) Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415: 451–454 [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku H, Takeda N, Charpentier M, Perry J, Miwa H, Umehara Y, Kouchi H, Murakami Y, Mulder L, Vickers K, et al. (2005) Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433: 527–531 [DOI] [PubMed] [Google Scholar]
- Ishikawa C, Hatanaka T, Misoo S, Fukayama H. (2009) Screening of high kcat Rubisco among Poaceae for improvement of photosynthetic CO2 assimilation in rice. Plant Prod Sci 12: 345–350 [Google Scholar]
- Ishikawa C, Hatanaka T, Misoo S, Miyake C, Fukayama H. (2011) Functional incorporation of sorghum small subunit increases the catalytic turnover rate of Rubisco in transgenic rice. Plant Physiol 156: 1603–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkehabadi S, Peddi SR, Anwaruzzaman M, Taylor TC, Cederlund A, Genkov T, Andersson I, Spreitzer RJ. (2005) Chimeric small subunits influence catalysis without causing global conformational changes in the crystal structure of ribulose-1,5-bisphosphate carboxylase/oxygenase. Biochemistry 44: 9851–9861 [DOI] [PubMed] [Google Scholar]
- Laisk A, Loreto F. (1996) Determining photosynthetic parameters from leaf CO2 exchange and chlorophyll fluorescence: ribulose-1,5-bisphosphate carboxylase/oxygenase specificity factor, dark respiration in the light, excitation distribution between photosystems, alternative electron transport rate, and mesophyll diffusion resistance. Plant Physiol 110: 903–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Sakashita H, Hidema J, Mae T, Ojima K, Osmond B. (1992) Distinctive responses of ribulose-1,5-bisphosphate carboxylase and carbonic anhydrase in wheat leaves to nitrogen nutrition and their possible relationships to CO2-transfer resistance. Plant Physiol 100: 1737–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Shimada T, Takumi S, Kaneko K, Matsuoka M, Shimamoto K, Nakano H, Miyao-Tokutomi M, Mae T, Yamamoto N. (1997) Does decrease in ribulose-1,5-bisphosphate carboxylase by antisense rbcS lead to a higher N-use efficiency of photosynthesis under conditions of saturating CO2 and light in rice plants? Plant Physiol 114: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto C, Miyazawa S, Ohkawa H, Fukuda T, Taniguchi Y, Murayama S, Kusano M, Saito K, Fukayama H, Miyao M. (2010) Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proc Natl Acad Sci USA 107: 5226–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Numazawa T. (1991) Cis-acting elements in the pyruvate, orthophosphate dikinase gene from maize. Mol Gen Genet 228: 143–152 [DOI] [PubMed] [Google Scholar]
- Parry MA, Andralojc PJ, Scales JC, Salvucci ME, Carmo-Silva AE, Alonso H, Whitney SM. (2013) Rubisco activity and regulation as targets for crop improvement. J Exp Bot 64: 717–730 [DOI] [PubMed] [Google Scholar]
- Peterhansel C, Offermann S. (2012) Re-engineering of carbon fixation in plants: challenges for plant biotechnology to improve yields in a high-CO2 world. Curr Opin Biotechnol 23: 204–208 [DOI] [PubMed] [Google Scholar]
- Peters JL, Silverthorne J. (1995) Organ-specific stability of two Lemna rbcS mRNAs is determined primarily in the nuclear compartment. Plant Cell 7: 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Rodermel S, Haley J, Jiang CZ, Tsai CH, Bogorad L. (1996) A mechanism for intergenomic integration: abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc Natl Acad Sci USA 93: 3881–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Schwender J, Ohlrogge JB. (2004) The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol 136: 2700–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. (2002) Variation in the kcat of Rubisco in C3 and C4 plants and some implications for photosynthetic performance at high and low temperature. J Exp Bot 53: 609–620 [DOI] [PubMed] [Google Scholar]
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y. (2004) Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432: 779–782 [DOI] [PubMed] [Google Scholar]
- Sharwood RE, von Caemmerer S, Maliga P, Whitney SM. (2008) The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiol 146: 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverthorne J, Tobin EM. (1990) Post-transcriptional regulation of organ-specific expression of individual rbcS mRNAs in Lemna gibba. Plant Cell 2: 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ. (2003) Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch Biochem Biophys 414: 141–149 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Makino A. (2012) Availability of Rubisco small subunit up-regulates the transcript levels of large subunit for stoichiometric assembly of its holoenzyme in rice. Plant Physiol 160: 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi K, Yoshizawa R, Mae T, Makino A. (2009) Differences in expression of the RBCS multigene family and Rubisco protein content in various rice plant tissues at different growth stages. Plant Cell Physiol 50: 1851–1855 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ohkubo M, Hatakeyama H, Ohashi K, Yoshizawa R, Kojima S, Hayakawa T, Yamaya T, Mae T, Makino A. (2007) Increased Rubisco content in transgenic rice transformed with the ‘sense’ rbcS gene. Plant Cell Physiol 48: 626–637 [DOI] [PubMed] [Google Scholar]
- Tada Y, Sakamoto M, Matsuoka M, Fujimura T. (1991) Expression of a monocot LHCP promoter in transgenic rice. EMBO J 10: 1803–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez GG, Farquhar GD, Andrews TJ. (2006) Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci USA 103: 7246–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vies SM, Bradley D, Gatenby AA. (1986) Assembly of cyanobacterial and higher plant ribulose bisphosphate carboxylase subunits into functional homologous and heterologous enzyme molecules in Escherichia coli. EMBO J 5: 2439–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer S, Quick WP (2000) Rubisco: physiology in vivo. In RC Leegood, TD Sharkey, S von Caemmerer, eds, Photosynthesis: Physiology and Metabolism. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 85–113 [Google Scholar]
- Waegemann K, Soll J. (1996) Phosphorylation of the transit sequence of chloroplast precursor proteins. J Biol Chem 271: 6545–6554 [DOI] [PubMed] [Google Scholar]
- Wang YL, Zhou JH, Wang YF, Bao JS, Chen HB. (2001) Properties of hybrid enzymes between Synechococcus large subunits and higher plant small subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase in Escherichia coli. Arch Biochem Biophys 396: 35–42 [DOI] [PubMed] [Google Scholar]
- Wanner LA, Gruissem W. (1991) Expression dynamics of the tomato rbcS gene family during development. Plant Cell 3: 1289–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmann CC, Ramage RT, Bohnert HJ, Ostrem JA. (1989) Identification of an assembly domain in the small subunit of ribulose-1,5-bisphosphate carboxylase. Proc Natl Acad Sci USA 86: 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Sharwood RE, Orr D, White SJ, Alonso H, Galmés J. (2011) Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) carboxylation rate in Flaveria. Proc Natl Acad Sci USA 108: 14688–14693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M, Putterill JJ, Ross GS, Laing WA. (2001) Determination of the relative expression levels of Rubisco small subunit genes in Arabidopsis by rapid amplification of cDNA ends. Anal Biochem 291: 237–244 [DOI] [PubMed] [Google Scholar]