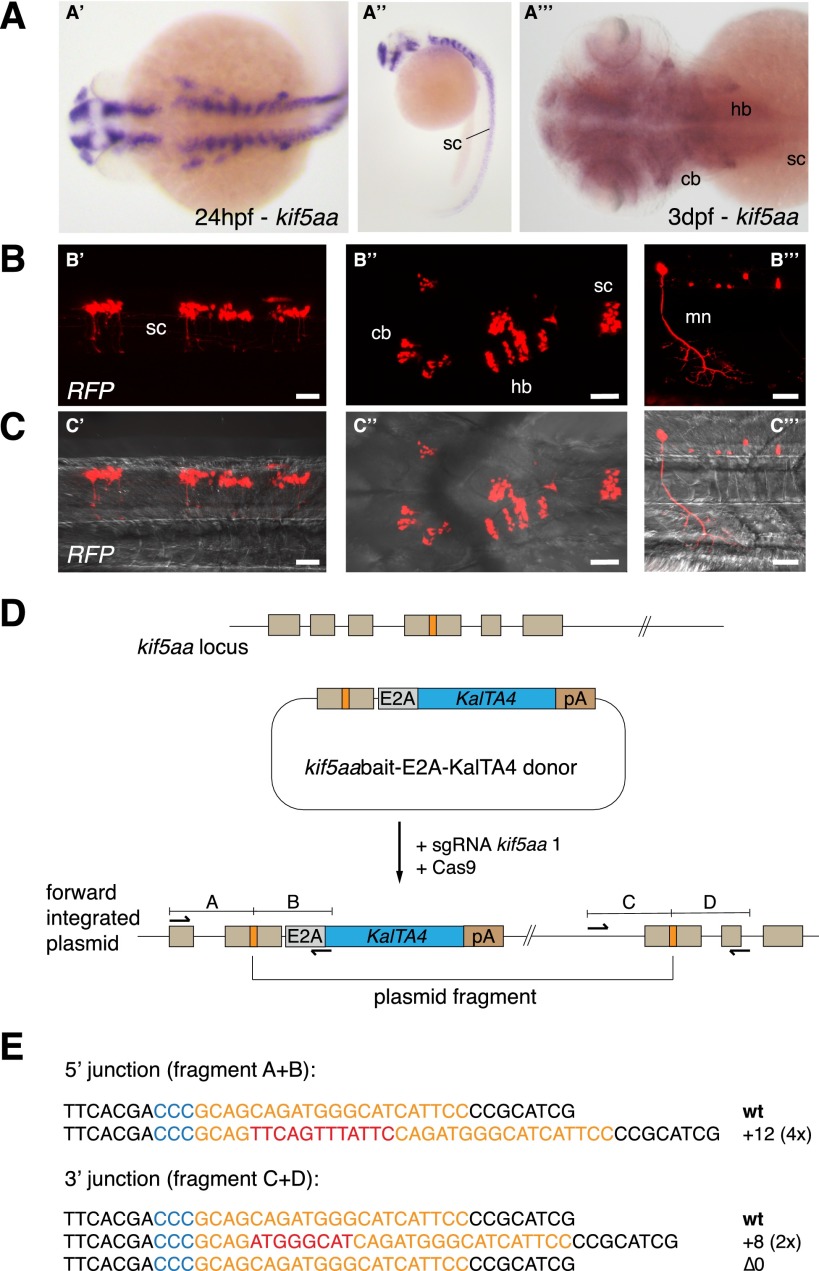
Figure 3.
CRISPR/Cas-mediated knock-in of KalTA4 into the kif5aa locus. (A) Kif5aa expression in zebrafish embryos revealed by in situ hybridization. Dorsal (A′) and lateral (A′′) views of 24-hpf embryos and dorsal view of 3-dpf embryo head and trunk region (A′′′) showing kif5aa expression in various brain regions and the spinal cord. (B,C) Representative confocal pictures of a Tg(UAS:RFP, cry1:eGFP) embryo showing RFP expression in the brain and spinal cord upon injection of the kif5aa bait donor plasmid together with sgRNA kif5aa 1 and Cas9 mRNA. Lateral view of the spinal cord (B′,C′), dorsal view of the head and trunk region (B′′,C′′), and high magnification of the spinal cord region (B′′′,C′′′) showing RFP expression in motoneurons. Scale bar, 50 μm. (sc) Spinal cord, (cb) cerebellum, (hb) hindbrain, (mn) motoneuron (cf. the GFP expression in the kif5aa BAC transgenic line reported by Kawasaki et al. [2012]). (D) A schematic of the used donor plasmid consisting of an N-terminal kif5aa bait with the sgRNA target site. The same E2A-KalTA4-pA cassette as in Figure 1A was used. (E) Sequence analysis at the 5′ and 3′ junctions of representative targeted integration events after PCR-based amplification. Binding sites of primers used for amplification are shown in D. (Orange) sgRNA binding site; (blue) PAM sequence NGG; (red) integrated additional base pairs. Note that the sgRNA is targeting the minus strand.