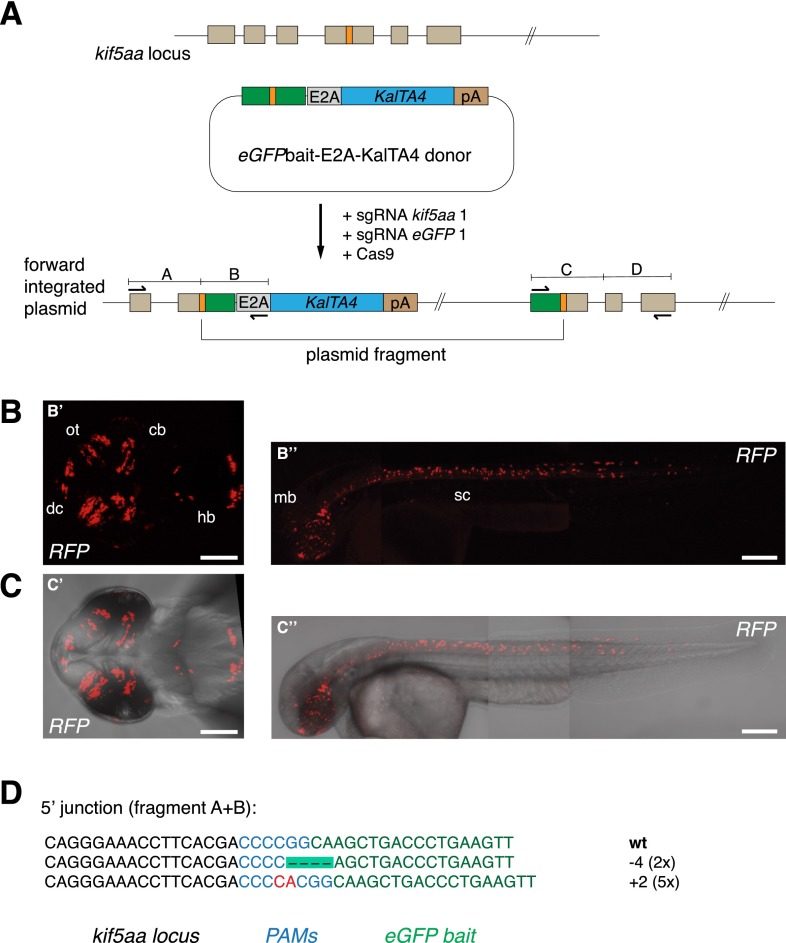
Figure 4.
CRISPR/Cas-mediated knock-in of KalTA4 into the kif5aa locus using the eGFPbait donor plasmid. (A) For integration of the E2A-KalTA4-pA cassette into the kif5aa locus, we used the eGFPbait donor plasmid in combination with two different sgRNAs. While sgRNA kif5aa 1 guides cleavage to the endogenous kif5aa locus, sgRNA eGFP 1 is employed for cleavage of the donor plasmid. (B,C) Representative confocal pictures of Tg(UAS:RFP, cry1:eGFP) 2-dpf embryos showing RFP expression in various brain regions and the spinal cord. Dorsal view (B′,C′) of the brain region and lateral view of an entire embryo (B′′,C′′) showing RFP expression in the whole length of the spinal cord and in the midbrain. Scale bar (B′,C′): 50 μm, (B′′,C′′): 200 μm. (dc) Diencephalon, (cb) cerebellum, (ot) optic tectum, (hb) hindbrain, (mb) midbrain, (sc) spinal cord. (D) Sequence analysis at the 5′ junction of representative targeted integration events after PCR-based amplification. Binding sites of primers used for amplification are shown in A. (Black) kif5aa locus; (blue) NGG PAM sequences for sgRNA kif5aa 1 and sgRNA eGFP 1; (green) parts of the eGFP bait sequence; (red) integrated additional base pairs. Note that, in this case, due to the frame difference between the kif5aa and eGFP genes, only +2 or −1 indels will produce functional fusion protein.