Abstract
Background:
Great gerbils, Rhombomys opimus, are the main reservoir host of zoonootic cutaneous leishmaniasis (ZCL) in Iran and neighboring countries. Based on morphological traits two subspecies R. opimus sodalis and R. opimus sargadensis have reported in the country. However, variation in infection rate and signs to Leishmania parasites, phenotype, size, and sexual polymorphisms demand more details to elucidate clearly the role of great gerbils in ZCL epidemiology.
Methods:
PCR-RFLP and PCR-direct sequencing were used to analyze mitochondrial DNA cytochrome B (mtDNA-cytB) gene structure of R. opimus collected from Golestan and Khorasan-e-Razavi Provinces in 2011 that are neighbor to Turkmenistan Country where ZCL is endemic in both sides of the borderline.
Results:
All of the specimens (n= 61) were morphologically or genetically similar to the typical R. opimus sodalis. However, there were 9 (1.5%) DNA substitutions throughout the 583 bp of the Cyt b gene of the samples sequenced comprising six DNA haplotypes. Maximum likelihood or neighbor joining phylogenetic trees inferred from the sequences could resolve the populations according to their subspecies as well as geographical origins.
Discussion:
The DNA polymorphisms in the great gerbils may correspond to the signs and infection rate in the animal. However, further studies are needed to match these six haplotypes with different signs and parasite sustaining following infection with L. major in the great gerbils.
Keywords: Leishmaniasis, ZCL, Rombomys opimus, mtDNA cytB, Iran
Introduction
Leishmaniasis is considered an emerging and re-emergent disease, with an increase in its incidence in the last decades (Reithinger et al. 2007). It has a global estimated prevalence of 12 million cases, with an estimative of 1.5–2 million new cases each year. At the present moment leishmaniasis occurs in 88 countries throughout Europe, Africa, Asia and America, and 350 million people are at risk of contracting the disease (Ameen 2010, WHO 2012). Ninety percent of annual cases are reported from Afghanistan, Brazil, Iran, Peru, Saudi Arabia, Syria, Algeria and Sudan (Desjeux 2004). Zoonotic Coutaneous leishmaniasis (ZCL) due to Leishmania major is one of the infectious increasing diseases in Iran where it almost doubled (from 11505 to 22705 cases) over a nine-year period in 2001 to 2009 (IMHME 2010, Oshaghi et al. 2010). Leishmaniasis is endemic in Turkmenistan which has a long borderline in northeast of Iran, and over the years 2000–2009, it has been reported 1,562 cases of CL, mostly at the southern territories bordering Iran (WHO 2009). Some of the most important causes of this upward trend are the movement of human populations toward the reservoir as well as vector habitats and the development of irrigation project.
The great gerbils, Rhombomys opimus, is known to be the most important reservoir host of L. major, being transmitted by sand flies of the genus Phlebotomus (Yaghoobi-Ershadi et al. 1996, Yaghoobi-Ershadi and Javadian 1996, Mohebali et al. 2004, Rassi et al. 2008a, 2011, Akhavan et al. 2010a,b,c, Oshaghi et al. 2008 and 2010). This rodent is distributed both in the arid and semi-arid regions throughout central and south Asia from Kazakhstan, Kyrgyzstan, Turkmenistan, Uzbekistan, Tajikistan, Iran, Pakistan, Afghanistan, to southern Mongolia, and north-western China (Sosnina 1979, Mallon 1985, Strelkova et al. 2001, Molur et al. 2005, Smith and Xie 2008, Shar et al. 2009, Abai et al. 2010). In Iran, it is widely distributed in central and northeast parts of the country (Yaghoobi-Ershadi and Javadian 1996, Rassi et al. 2008b, Akhavan et al. 2010a,b,c, Oshaghi et al. 2011).
The rate of infection of R. opimus to L. major is normally high and may reach to 92.5% at endemic areas (Rassi et al. 2008a). The parasite persists in the great gerbils for up to 25 months (Strelkova et al. 2001). They are also known to carry the germs of bubonic plague (Davis et al. 2004, Zhang and Dai 2008) as well as being an agricultural pest (Nowak 1999).
The great gerbils occupy desert to semi-desert habitats, and are associated with succulent plants of family Chenopodiaceae (Climacoptera spp., Salsola spp., Suaeda spp.) (Dubrovsky 1975, Rogovin et al. 2003). These saline ecological biotopes are discontinuous in distribution in center, north, and northwest of Iran, thus leading to a fragmentation of the populations of great gerbils. Based on morphometric characteristics and phenotypic differences, four subspecies namely R. opimus opimus, R. opimus furmicolor, R. opimus sargadensis, and R. opimus sodalis have been reported within the species (Goodwin 1940, Etemad 1978). Two subspecies of R. opimus sargadensis and R. opimus sodalis occur in Iran (Abai et al. 2010). R. opimus sodalis is bigger than R. opimus sargadensis. This subspecies is brown to hazelnut color, whereas R. opimus sargadensis has a yellowish color. These phenotypic variations are related to the wide geographic distribution of the great gerbils. Additionally, sexual dimorphism and variation in the size of different developmental stages may make the diagnosis of the subspecies more challenging.
Accurate identification of reservoir host species is important in the epidemiology and control of zoonotic diseases such as ZCL (Meerburg et al. 2009, Sakthianandeswaren et al. 2009). Although great gerbils have a fundamental role in the epidemiology of ZCL, not many studies have been focused on the relationship of R. opimus population structure and ZCL prevalence or severity of the disease. It is shown a considerable variation in infection rate and signs to Leishmania parasites. For example the rate of Leishmania infection and its signs ranged from 11–92.5% and asymptotic to scar respectively among different R. opimus populations in Iran (Yaghoobi-Ershadi et al. 1996, Mohebali et al. 2004, Rassi et al. 2008a, Akhavan et al. 2010b,c). In another study, it was shown that exposure of R. opimus populations to L. major resulted a wide range of reactions, out of 194 gerbils examined, L. major and L. turranica were isolated from 80 (41.23%) of the animals, 55 (68.75%) of the infected animals showed no skin lesion (asymptomatic), and only 25 (31.25%) of the animals showed skin lesions and even the rate of parasitemia was not uniform (Hajjaran et al. 2013). This seems to be due to various physiological and genetic traits that affect the host-parasite interaction. Therefore, studying the genetic structure of different R. opimus populations is of fundamental importance in ZCL epidemiological investigation. At least two major lineages (subspecies) exist within the species of R. opimus in Iran and analysis of the mitochondrial cytochrome b (mtDNA-cytB) gene, revealed a range of 1–10% genetic variation among the populations of seven districts belonging to five provinces in central and north-eastern Iran (Oshaghi et al. 2011).
In the current study, we aimed to characterize the mtDNA-CyB gene sequences of three great gerbil populations distributed in northeastern Iran close to the border line with Turkmenistan where ZCL is endemic.
Materials and Methods
Study area and rodent collection
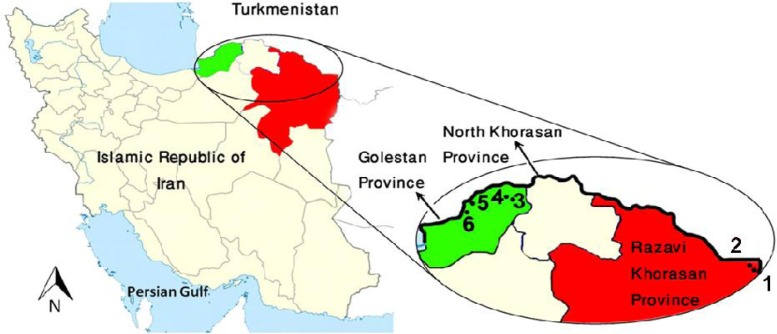
This study was conducted in three districts of northeastern part of Iran including Sarakhs of Khorassan-e-Razavi Province, and Gonbad-e Qabus and Maraveh-Tappeh from Golestan Province in 2011. Details of collection sites are shown in Table 1. These districts respectively are located in the northern and far northeastern parts of the borderline of Iran-Turkmenistan (Fig. 1).
Table 1.
Details of Rhombomys opimus specimens collected from the area study close to borderline of Iran-Turkmenistan
| Subspecies | Province (Country) | County | Village(s) | No. | Latitude–Longitude | Genbank A.N. |
|---|---|---|---|---|---|---|
| R. opimus sodalis | Golestan (Iran) | Gonbad-e-Qabus | Dashli-Borun, Fadavi | 5 6 |
37.3–54.5 37. 3–54.4 |
JX412206 JX412213-4 |
| R. opimus sodalis | Golestan (Iran) | Maraveh-Tappeh | Ghare Gol, Sozesh | 6 6 |
37.5–55.3 37.5–55.4 |
JX412207 JX412212 |
| R. opimus sodalis | Razavi Khorasan (Iran) | Sarakhs | Gonbadli, Sangar | 22 16 |
36.2–60.5 36.1–61.1 |
JX412210-1 JX412215-6 JX412208-9 |
| R. opimus sodalis | Golestan (Iran) | Kalaleh | Qareh Gol Gharbi | - | 37.9–55.7 | FJ648772* |
| R. opimus sodalist/ sargadensis Hybrid | Northern-Khorasan (Iran) | Kalaleh | Qareh Gol Gharbi | - | 37.9–55.7 | FJ648775* |
| R. opimus NI | Iran | NI | NI | - | NI | AJ430556* |
| R. opimus sargadensis | Northern-Khorasan (Iran) | Esfarayen | Kalateh-Shur | - | 37.0–57.1 | FJ648773* |
| R. opimus sargadensis | Northern-Khorasan (Iran) | Shirvan | Hossein-abad | - | 37.2–57.7 | FJ648774* |
| R. opimus sargadensis | Semnan (Iran) | Shahrood | Ahmadabad Mayamey |
- | 35.4–52.2 36.4–55.6 |
FJ648771* FJ648770* |
| R. opimus sargadensis | Semnan (Iran) | Damghan | Soltanieh | - | 36.0–54.3 | FJ648769* |
| R. opimus sargadensis | Esfahan (Iran) | Kashan | Badrood | - | 32.6–52.0 | FJ648768* |
| R. opimus sargadensis | Esfahan (Iran) | Habibabad | Habibabad | - | 33.3–52.5 | FJ648767* |
| Meriones lybicus | Xinjiang (China) | Uygur | Turfan | - | 42.5–89.1 | AB38190* |
The asterisk (*) show the specimens obtained from genbank database. NI: Not indicated
Fig. 1.
Study areas and the location where Rhombomys opimus specimens were collected. 1, Sangar, 2, Gonbadli, 3, Sozesh, 4, Ghareh Gol, 5, Dashli-Borun, and 6, Fadavi
Animal collections were performed using Sherman live-trap (30cm× 15cm× 15cm wire mesh) baited with a mixture of walnut, cucumber, tomato, and bread dabbed with sunflower oil. Approximately 30–40 live traps were used per night at each location in monthly collections. The traps were set close to burrow entrance 2–3 h before the dusk and were checked in the next morning after sunrise. The captured animals were transported to the laboratory, anesthetized using a mixture of 150mg/kg ketamine 10% and 15mg/kg xylazine 2% before 200–1000μL of blood was taken from the tail vein in tubes containing 20–30μL anticoagulants (heparin, EDTA, 0.29 M) and stored at −20 °C until use. We also used DBC (DNA Banking Card) to store blood samples of the rodents. Blood sample (50μL) were spotted directly on separate DBC cards and dried at room temperature as explained previously by Karimian et al. (2011). Morphological measurements were also recorded while the animals were anesthetized.
All experiments on the rodents were performed in accord with the guidelines of the Ethical Board of Tehran University of Medical Sciences, Iran.
Species and subspecies identification
The genus and species of the rodents were determined by external morphological characteristics including color, grooves on the incisor teeth, length of ears, tail, hind feet, head, body, and skull (Ziaei 1996). Presence of two fine grooves on the incisor teeth is the most important characteristics for R. opimus identification. The specimens with hazel-nut color, body length ranging from 300 to 320mm and a tail length of 135 to 140mm, and hind feet length of 43 to 45mm were considered to be R. opimus sodalis subspecies. Specimens with brown to yellowish color, body length ranging from 200 to 250mm and a tail length of 100 to 130mm, were determined to be R. opimus sargadensis subspecies (Abai et al. 2010).
mtDNA cytB PCR and sequencing
One mm diameter disc of each DBC card containing dried blood sample were punched and then DNA was extracted according to the manufacturer’s guide (Karimian et al. 2011) and used for PCR amplification. We also extracted DNA from the blood samples (200μL blood per animal) using G-spin Blood and Tissue DNA Kit (Bioneer, South Korea) according to the manufacturer’s instructions.
A 624bp fragment of the cytochrome b gene (cytB) of the mitochondrial (mt) DNA were amplified using the protocol of (Maleki-Ravasan et al. 2009) and the primers described by Kent and Norris (Kent and Norris 2005). They primers used were UNFOR403 (5′-T GAGGACAAATATCATTCTGAGG-3′) and UNREV1025 (5′-GGTTGTCCTCCAATTCA TGTTA-3′). The PCR amplification was performed in 25μL reaction mixtures containing 10mM Tris, pH 8.3, 50mM KCl, 1.5mM Mg Cl2, 0.01% gelatin, 1.0mM deoxynucleotide triphosphates, 1.25 units of Taq polymerase, 50pmol of each primer, and 5μL of the extracted DNA or and one mm disk of DBC card. An initial denaturation step at 95 °C for 5min was applied prior to 35 cycles of denaturation for 1 min at 95 °C, hybridization for 1 min at 58 °C and extension for 1 min at 72 °C followed by a final extension at 72 °C for 7min. Some DNA of R. opimus sodalis (Oshaghi et al. 2011) were prepared from the Insect Molecular Biology lab at the School of Public Health and used as positive controls in PCR and PCR-RFLP analysis.
PCR products were analyzed by electrophoresis on 1% agarose gel stained with ethidium bromide. A subset of PCR products, representatives of populations of the three districts were selected for sequencing (Table 1). Sequencing was performed using an ABI 3730 sequencer machine by Seqlab (Göttingen, Germany).
Sequences were checked to correct ambiguities. Homologies with the available sequence data in GenBank was checked using basic local alignment search tool (BLAST) analysis software (www.ncbi.nlm.nih.gov/BLAST). The cytB gene sequences of the great gerbil R. opimus available in genbank database (Table 1) were retrieved and used as reference to generate multiple alignments using the Clustal W software (Higgins and Sharp 1988) and phylogenetic analysis.
PCR-RFLP and phylogenetic analysis
The sequences of the cytB gene of the great gerbils obtained in this study were checked by the specific restriction enzymes previously introduced by (Oshaghi et al. 2011). HinfI restriction enzymes (RE) could identify subspecies of R. opimus sodalis from R. opimus sargadensis. Digestions were performed in 25μL mixtures containing 15μL of PCR product mixed with 2.5μL of the enzyme buffer and 5 units of HinfI RE overlaid with two drops of mineral oil. The mixture was incubated at the temperature recommended by the enzyme suppliers. An aliquot (14μL) of the digestion product was mixed with 6μL of loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol, and 30% glycerol), and electrophoresed in a 2.5% agarose gel. The gels were stained with ethidium bromide (2mg/mL) and the RFLP profiles were visualized under ultraviolet light.
Phylogenetic analysis was performed by comparing the sequences obtained in this study plus the ones were available in Genbak for R. opimus cytB gene. Neighbor joining (NJ) and maximum parsimony (MP) trees were constructed by the algorithm in MEGA 4.0 (Tamura et al. 2007). Robustness of the phylogenetic trees was tested with bootstrapping value. CytB sequence of Meriones lybicus (AB381902) was acquired from Genbank and used as an out group in this study (Table 1).
Results
Rhombomys opimus subspecies
A total of 61 great gerbil specimens including 25 females and 36 males were collected in this study. Details of the captured specimens are shown in Table 1. Approximately 68 percent (n= 38) of the rodents were caught from Sarakhs District. Field observation revealed that number of rodent burrows in Gonbadli village was twice as other villages.
The genus and species of the rodents were determined by testing external morphological characteristics already explained in M and M section. Comparison between the populations was performed for specimens of the same sex. According to the classification mentioned above, we could find just one subspecies R. opimus sodalis in the study area. All of the specimens had a color like hazel-nut and the range of body length was 120–180mm without calculating tail length. The average of the body length without calculating the tail was 163mm. This is important that some of these rodents may not be adult and so their body length may be shorter than adult ones. Weights of these rodents were ranged from 112 to 269 grams and the average weight was 176 grams.
CytB sequence and PCR-RFLP
The sequences of mtDNA cytB gene of all 61 R. opimus specimens captured in this study were analyzed by either sequencing or PCR-RFLP assays. The PCR amplicons of all specimens showed a single band of the expected size (624bp) on the agarose gel. Eleven specimens, as representatives of the populations or villages (Table 1) were selected for sequencing. Almost 583–586bp of results of sequencing were trustable for analysis. We found six mitochondrial cytB haplotypes among the specimens sequenced (Table 2). When we added other R. opimus sodalis sequences available in genbank, totally eight haplotypes were found in the subspecies (Table 2). There was a strong correlation between the haplotypes and their geographical origins.
Table 2.
DNA sequence comparison of about 583 bp of CytB gene of Rhombomys opimus sodalis populations distributed in northeastern Iran
| Origin | Haplotype | Accession number |
Position in the PCR product
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 1 | 1 | 2 | 3 | 3 | 3 | 4 | 4 | 4 | 5 | |||
| 6 | 8 | 5 | 6 | 6 | 4 | 6 | 8 | 2 | 6 | 7 | 4 | |||
| 3 | 1 | 6 | 9 | 7 | 0 | 5 | 4 | 3 | 5 | 7 | 9 | |||
| Gonbadli, Sarakhs | I | JX412210 | C | G | T | C | C | T | C | C | C | C | A | A |
| Gonbadli, Sarakhs | I | JX412211 | . | . | . | . | . | . | . | . | . | . | . | . |
| Gonbadli, Sarakhs | I | JX412215 | . | . | . | . | . | . | . | . | . | . | . | . |
| Fadavi, Golestan | II | JX412214 | . | . | C | . | . | . | . | . | . | . | . | . |
| Ghare Gol, Golestan | II | JX412207 | . | . | C | . | . | . | . | . | . | . | . | . |
| Gonbadli, Sarakhs | III | JX412216 | . | . | C | . | T | . | . | . | . | . | . | . |
| Dashli-Borun, Golestan | IV | JX412206 | . | A | C | . | . | . | . | . | . | . | . | . |
| Sangar, Sarakhs | IV | JX412209 | . | A | C | . | . | . | . | . | . | . | . | . |
| Sangar, Sarakhs | IV | JX412208 | . | A | C | . | . | . | . | . | . | . | . | . |
| Sozesh, Golestan | V | JX412213 | . | . | C | T | . | . | . | T | T | . | G | . |
| Fadavi, Golestan Kalaleh, Golestan | VI | JX412212 | T | . | C | . | . | . | T | A | T | . | G | . |
| Shahrood/Kalaleh | VII | FJ648772 | # | . | C | . | . | G | T | T | T | . | G | . |
| Iran | VIII | FJ648775 | . | . | C | . | . | . | . | . | . | T | . | G |
| IX | AJ430556 | . | A | C | . | . | . | . | . | . | . | . | . | |
Dots show identical sequences to the top sequence. Three last row sequences derived from genbank (Accession no. FJ648772 and FJ648775, Oshaghi et al. 2011, AJ430556, Chevre 2005: direct submission).
Not indicated
Generally, the sequences were more or less similar to the sequences of typical R. opimus sodalis (Accession Nos: FJ648772, FJ648775, and AJ430556) previously reported in Iran. However, there were 9 (1.5%) DNA substitutions throughout the 583bp of the Cyt b gene of the samples sequenced. When other R. opimus sodalis sequences added to the analysis the number of substitutions rose to 12. Most of the nucleotide substitutions (92%) were transition and, except one amino acid (AA) substitution, there were no AA substitution throughout the 194 base of AA in the cytB region of the populations.
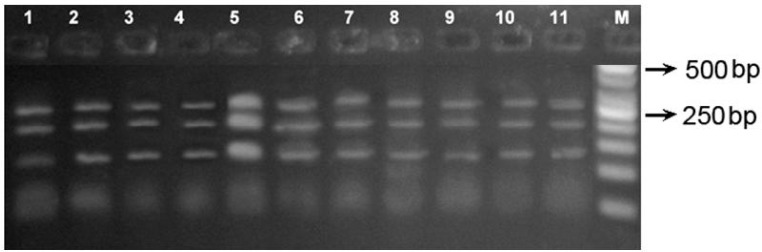
HinfI restriction enzyme showed three bands of 284, 212, and 128bp which is a specific RFLP profile for R. opimus sodalis for all of the 61 specimens as well as the R. opimus sodalis from Kalaleh population as positive control (Fig. 2).
Fig. 2.
PCR-RFLP profiles (284, 212, and 128 bp) of CytB gene for Rhombomys opimus sodalis digested by HinfI RE. All the specimens were identified as R. opimus sodalis. M: 100 bp ladder (Sinaclone, Iran). Specimens in lanes 1–2: Sangar (Sarakhs), 3–4: Dashli-Borun (Gonbad-e-Qabus), 5: reference strain of R. opimus sodalis from Kalaleh (Gorgan), 6–7: Gonbadli (Sarakhs), 8: Ghare-Gol (Maraveh-Tappeh), 9: Sozesh (Maraveh-Tappeh), and 10–11 Fadavi (Gonbad-e-Qabus)
Phylogenetic analysis
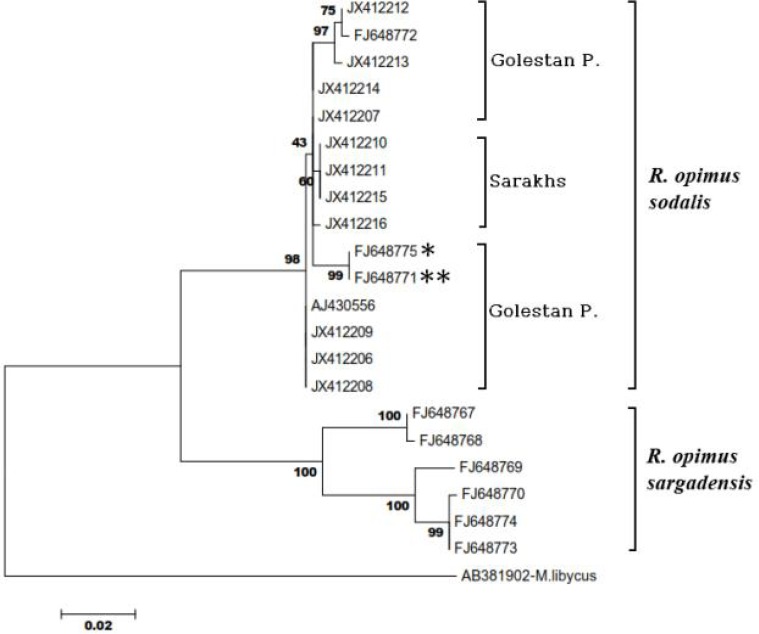
Tree reconstruction on the combination of data obtained in this study plus those from genbank using neighbor-joining (NJ) and maximum parsimony (MP) methods revealed similar topology for the R. opimus populations or subspecies. The trees showed two main clades resolving very well the R. opimus sodalis from R. opimus sargadensis (Fig. 3). Specimens of Gonbad-e-Qabus, Maraveh-Tappeh, and Sarakhs districts were associated with the specimen of Kalaleh district and the ones already known as R. opimus sodalis, altogether grouped in one clade whereas the second group included the R. opimus sargadensis (Fig. 3). Within the clade of R. opimus sodalis three major branches were observed, two of them contained specimens of Golestan and the other one included the Saraks specimens. The specimens of Golestan were scattered in two distinct subclades. Sarakhs specimens were positioned in between Golestan specimens. Except three cases, the bootstrap values of the nodes were more than 90 percent indicating high confidence in tree topology and well resolutions between the populations or subspecies.
Fig. 3.
Phylogenetic relationship between Rhomomys opimus subspecies/populations of Iran based on sequence analysis of 583 bp mtDNAcytB gene. Sequences this study (JX412206-16) was combined with the available sequences from the Genbank database (Table 1). Species of Meriones lybicus (AB381902, Ito, direct submission) was used as an outgroup. *is a hybrid specimen of R. opimus sodalis from Golestan (female) with R. opimus sargadensis (male) from Shahrood which showed maternal sodalis haplotype. **was a specimen migrated from Golestan to Shahrood (Oshaghi et al. 2011). Genetic distance scale is shown underneath
Discussion
In this study, a combination of morphological and molecular (PCR-RFLP, PCR-direct sequencing, phylogenetic topology) characteristics were used to reveal taxonomic situation and genetic structure of the R. opimus populations scattered in the north and northeast of Iran where ZCL is endemic and the rodents act as the main reservoir host of the disease. This investigation revealed the presence of six haplotypes, all were of R. opimus sodalis subspecies in the study area. This finding is in agreement with the previous report indicating the presence of R. opimus sodalis in Kalaleh district, an adjacent area in Golestan Province in north of Iran (Oshaghi et al. 2011). We have no access to the populations of great gerbils of Turkmenistan on the other side of border line with Iran; however, due to presence of similar ecological niches in those areas we suggest the presence of similar subspecies or haplotype in Turkmenistan. We suggest testing the genetic structure of great gerbils which is known as the main host of Leishmania parasites causing ZCL in the neighboring country.
All of the areas investigated in this research comprising Gonbad-e-Qabus, Maraveh-Tappeh, and Sarakhs districts plus Kalaleh district are located in northern slopes of Alborz Mountain Chain (AMC) (Fig. 1). The AMC in north of Iran acts as a natural geographical barrier between two main subspecies and seems to be the southern limit of R. opimus sodalis in the region. This natural barrier between R. opimus sargadensis and R. opimus sodalis resulted in accumulation of mutations, independent evolution, and fragmentation between the great gerbil populations/ subspecies. Considering presence of 2–10 percent intra-specific genetic variation among the populations (Oshaghi et al. 2011) may lead to speciation in future if gene flow does not occur between them.
Phylogenetic analysis revealed good resolution between the populations as well as subspecies. However, presence of high rate of sequence homology between a few specimens originated from two distinct areas (Sarakhs and Golestan) suggested possible transportation via vehicles (train, bus, and car) or natural migration from Sarakhs to Golestan Province and vise versa. The distance between these two locations is about 500Km, however the areas in between are plain and there is no significant natural barrier to obstruct merging these populations and transportation could have been happened easily throughout the time by this active animal. This observable fact can also be explained by present of a common ancestor in those regions. This phenomenon also has been observed in the previous study indicating possible migration of R. opimus sodalis from Kalaeh to the territory of R. opimus sargadensis in Shahrood district (Oshaghi et al. 2011).
Different populations of great gerbils have different rate of Leishmania infection which could affect their ability to sustain L. major (YaghoobiErshadi et al. 1996, Mohebali et al. 2004, Rassi et al. 2008a,b, Akhavan et al. 2010b). Leishmaniasis clinical manifestation depends upon Leishmania species and host genetic background which governs generation of type of immune response (Mohamed et al. 2003, Salhi et al. 2008, Sakthianandeswaren et al. 2009 and 2010, Castellucci et al. 2012, Moravej et al. 2012). Variation in maintaining pathogens such as plague bacilli or Rift Valley Fever Virus between populations of a given rodent species have been reported in literature (Korobitsyna 1974, Arntzen et al. 1991, Gora et al. 2000, Mills 2005).
In this study we have tested the mtDNA cytB gene between great gerbil populations and found 1.5% variation between them. However, supplementary studies are needed to assess whether or not these six haplotypes are correlated to various outcoms (symtomatic to asymtomatic) and to the ability L. major parasite to infect and to remain within the great gerbils. Also it is suggested testing other genes such as TNF-alpha, TNF-beta, IL-4, IL-10 and IFN-γ among R. opimus populations. These genes could influence the host-pathogen associations and could determine the severity of the disease upon infection with L. major (Kamali-Sarvestani et al. 2006, Salhi et al. 2008, Sakthianandeswaren et al. 2009 and references herein). These studies could be used to develop accurate ecological control strategies and provide important step towards deeper understanding of ZCL epidemiology particularly the host-pathogen associations among R. opimus populations.
Acknowledgments
This research was supported by research deputy of Tehran University of Medical Sciences. We are very grateful to Mr Naroyi and Mr Koohkan, the staff of Sangar, Gonbadli, Nobonyad and Sarakhs County health center for their helps in the field. We are also thanks Mr A Ataei, B Shiravand, M Hosseini, M Anjomruz and Mrs Kamran. The authors declare that there is no conflict of interest.
References
- Abai MR, Oshaghi MA, Tajedin L, Rassi Y, Akhavan AA. Geographical distribution and ecological features of the great gerbil subspecies in the main zoonotic cutaneous leishmaniasis foci in Iran. Asian Pac J Trop Med. 2010;3:800–803. [Google Scholar]
- Akhavan AA, Mirhendi H, Khamesipour A, Alimohammadian MH, Rassi Y, Bates P, Kamhawi S, Valenzuela JG, Arandian MH, Abdoli H, Jalali-zand N, Jafari R, Shareghi N, Ghanei M, Yaghoobi-Ershadi MR. Leishmania species: Detection and identification by nested PCR assay from skin samples of rodent reservoirs. Exp Parasitol. 2010a;126:552–556. doi: 10.1016/j.exppara.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhavan AA, Yaghoobi-Ershadi MR, Khamesipour A, Mirhendi H, Alimohammadian MH, Rassi Y, Arandian MH, Jafari R, Abdoli H, Shareghi N, Ghanei M, Jalali-Zand N. Dynamics of Leishmania infection rates in Rhombomys opimus (Rodentia: Gerbillinae) population of an endemic focus of zoonotic cutaneous leishmaniasis in Iran. Bull Soc Pathol Exot. 2010b;103:84–89. doi: 10.1007/s13149-010-0044-1. [DOI] [PubMed] [Google Scholar]
- Akhavan AA, Yaghoobi-Ershadi MR, Mirhendi H, Alimohammadian MH, Rassi Y, Shareghi N, Jafari R, Arandian MH, Abdoli H, Ghanei M, Jalali-Zand N, Khamesipour A. Molecular epizootiology of rodent leishmaniasis in a hyperendemic area of Iran. Iranian J Publ Health. 2010c;39:1–7. [PMC free article] [PubMed] [Google Scholar]
- Ameen M. Cutaneous leishmaniasis: Advances in disease pathogenesis, diagnostics and therapeutics. Clin Exp Dermatol. 2010;35:699–705. doi: 10.1111/j.1365-2230.2010.03851.x. [DOI] [PubMed] [Google Scholar]
- Arntzen L, Wadee AA, Isaäcson M. Immune responses of two Mastomys sibling species to Yersinia pestis. Infect Immun. 1991;59:1966–1971. doi: 10.1128/iai.59.6.1966-1971.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci L, Jamieson SE, Almeida L, Oliveira J, Guimarães LH, Lessa M, Fakiola M, Jesus AR, Nancy Miller E, Carvalho EM, Blackwell JM. Wound healing genes and susceptibility to cutaneous leishmaniasis in Brazil. Infect Genet Evol. 2012;12(5):1102–1110. doi: 10.1016/j.meegid.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Begon M, De Bruyn L, Ageyev VS, Klassovskiy NL, Pole SB, Viljugrein H, Stenseth NC, Leirs H. Predictive thresholds for plague in Kazakhstan. Science. 2004;304:736–738. doi: 10.1126/science.1095854. [DOI] [PubMed] [Google Scholar]
- Desjeux P. Leishmaniasis: Current situation and new perspectives. Comp Immunol Microbiol Infect Dis. 2004;27:305–318. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Dubrovsky YA. Ecological causes of predominance of some mammals as, reservoirs of Leishmania tropica major in Turanian deserts. Folia Parasitol. 1975;22:163–169. [PubMed] [Google Scholar]
- Etemad A. Publication of National Association of natural resources and human environment protection. first ed. Tehran University Publication; Tehran: 1978. Iranian mammals: rodents and their identification keys. [Google Scholar]
- Goodwin GG. Mammals collected by the Legendre 1938 Iran expedition. Am Mus Novit. 1940;1082:1–17. [Google Scholar]
- Gora D, Yaya T, Jocelyn T, Didier F, Maoulouth D, Amadou S, Ruel TD, Gonzalez JP. The potential role of rodents in the enzootic cycle of rift valley fever virus in Senegal. Microbes Infect. 2000;2:343–346. doi: 10.1016/s1286-4579(00)00334-8. [DOI] [PubMed] [Google Scholar]
- Hajjaran H, Mohebali M, Abaei MR, Oshaghi MA, Zarei Z, Charehdara S, Mirjalali H, Sharifdini M, Teimouri A. Natural infection and phylogenetic classification of Leishmania spp. infecting Rhombomys opimus, a primary reservoir host of zoonotic cutaneous leishmaniasis in northeast Iran. Trans R Soc Trop Med Hyg. 2013;107(9):550–557. doi: 10.1093/trstmh/trt060. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- IMHME IMHME: Iran Ministry of Health and Medical Education, 2010. 2010. Official Report of Leishmania Cases in Iran.
- Kamali-Sarvestani E, Rasouli M, Mortazavi H, Gharesi-Fard B. Cytokine gene polymorphisms and susceptibility to cutaneous leishmaniasis in Iranian patients. Cytokine. 2006;35:159–165. doi: 10.1016/j.cyto.2006.07.016. [DOI] [PubMed] [Google Scholar]
- Karimian F, Sedaghat MM, Oshaghi MA, Mohtarami F, Sanei Dehkordi A, Koosha M, Akbari S, Hashemi-Aghdam SS. Utility of filter paper for preserving insects, bacteria, and host reservoir DNA for molecular testing. Iran J Arthropod-Borne Dis. 2011;5(2):42–50. [PMC free article] [PubMed] [Google Scholar]
- Kent RJ, Norris DE. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome B. Am J Trop Med Hyg. 2005;73:336–342. [PMC free article] [PubMed] [Google Scholar]
- Korobitsyna KV. Analysis of the interpopulation karyotype variability in Meriones meridianus with special reference to the resistance against the plague microbe. Zool Zh. 1974;53:1066–1069. [Google Scholar]
- Maleki-Ravasan N, Oshaghi MA, Javadian E, Rassi Y, Sadraei J, Mohtarami F. Blood meal identification in field-captured sand flies: comparison of PCR-RFLP and ELISA assays. Iran J Arthropod-Borne Dis. 2009;3:8–18. [PMC free article] [PubMed] [Google Scholar]
- Mallon DP. The mammals of the Mongolian People’s Republic. Mammal Rev. 1985;15:71–102. [Google Scholar]
- Meerburg BG, Singleton GR, Kijlstra A. Rodent-borne diseases and their, risks for public health. Crit Rev Microbiol. 2009;35:221–270. doi: 10.1080/10408410902989837. [DOI] [PubMed] [Google Scholar]
- Mills JN. Regulation of rodent-borne viruses in the natural host: implications for human disease. Infectious Diseases from Nature: Mechanisms of Viral Emergence and Persistence. 2005:45–57. doi: 10.1007/3-211-29981-5_5. [DOI] [PubMed] [Google Scholar]
- Mohamed HS, Ibrahim ME, Miller EN, Peacock CS, Khalil EA, Cordell HJ, Howson JM, El Hassan AM, Bereir RE, Blackwell JM. Genetic susceptibility to visceral leishmaniasis in The Sudan: linkage and association with IL4 and IFNGR1. Genes Immun. 2003;4(5):351–355. doi: 10.1038/sj.gene.6363977. [DOI] [PubMed] [Google Scholar]
- Mohebali M, Javadian E, Yaghoobi-Ershadi MR, Akhavan AA, Hajjaran H, Abai MR. Characterization of Leishmania infection in rodents from endemic areas of the Islamic Republic of Iran. East Mediterr Health J. 2004;10:591–599. [PubMed] [Google Scholar]
- Molur S, Srinivasulu C, Srinivasulu B, Walker S, Nameer PO, Ravikumar L. Status of Non-Volant Small Mammals. Conservation Assessment and Management Plan (CAMP) Workshop Report; Coimbatore, India: 2005. [Google Scholar]
- Moravej A, Rasouli M, Kalani M, Asaei S, Kiany S, Najafipour S, Koohpayeh A, Abdollahi A. IL-1β (-511T/C) gene polymorphism not IL-1β (+3953T/ C) and LT-α (+252A/G) gene variants confers susceptibility to visceral leishmaniasis. Mol Biol Rep. 2012;39(6):6907–6914. doi: 10.1007/s11033-012-1517-z. [DOI] [PubMed] [Google Scholar]
- Nowak R. Walkers Mammals of the World. sixth ed. Johns Hopkins University Press; Baltimore: 1999. [Google Scholar]
- Oshaghi MA, Rassi Y, Tajedin L, Abai MR, Akhavan AA, Enayati A, Mohtarami F. Mitochondrial DNA diversity in the populations of great gerbils, Rhombomys opimus, the main reservoir of cutaneous leishmaniasis. Acta Trop. 2011;119:165–171. doi: 10.1016/j.actatropica.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Oshaghi MA, Rasolian M, Shirzadi MR, Mohtarami F, Doosti S. First report on isolation of Leishmania tropica from sandflies of a classical urban cutaneous leishmaniasis focus in southern Iran. Exp Parasitol. 2010;126(4):445–450. doi: 10.1016/j.exppara.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Oshaghi MA, Yaghobi-Ershadi MR, Abbassi M, Parvizi P, Akhavan AR, Foroshani AR, Zahraei AR, Rassi Y, Mohtarami F. Detection of Leishmania major in naturally infected sand flies using semi Nested-PCR. Iranian J Publ Health. 2008;37:59–64. [Google Scholar]
- Rassi Y, Oshaghi MA, Azani SM, Abaie MR, Rafizadeh S, Mohebai M, Mohtarami F, Zeinali Mk. Molecular detection of Leishmania infection due to Leishmania major and Leishmania turanica in the vectors and reservoir host in Iran. Vector Borne Zoonotic Dis. 2011;11(2):145–150. doi: 10.1089/vbz.2009.0167. [DOI] [PubMed] [Google Scholar]
- Rassi Y, Abai MR, Javadian E, Rafizadeh S, Imamian H, Mohebali M. Molecular data on vectors and reservoir hosts of zoonotic cutaneous leishmaniasis in central Iran. Bull Soc Pathol Exot. 2008a;101:425–428. [PubMed] [Google Scholar]
- Rassi Y, Sofizadeh A, Abai MR, Oshaghi MA, Rafizadeh S, Mohebail M, Mohtarami F, Salahi R. Molecular Detection of Leishmania major in the Vectors and Reservoir Hosts of Cutaneous Leishmaniasis in Kalaleh District, Golestan Province, Iran. Iran J Arthropod Borne Dis. 2008b;2:21–27. [Google Scholar]
- Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S. Cutaneous leishmaniasis. Lancet Infect Dis. 2007;7:581–596. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- Rogovin K, Randall JA, Kolosova I, Moshkin M. Social correlates of stress, in adult males of the great gerbil. Rhombomys opimus, in years of high and low, population densities. Horm Behav. 2003;43:132–139. doi: 10.1016/s0018-506x(02)00028-4. [DOI] [PubMed] [Google Scholar]
- Sakthianandeswaren A, Foote SJ, Handman E. The role of host genetics in, leishmaniasis. Trends Parasitol. 2009;25:383–391. doi: 10.1016/j.pt.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Sakthianandeswaren A, Curtis JM, Elso C, Kumar B, Baldwin TM, Lopaticki S, Kedzierski L, Smyth GK, Foote SJ, Handman E. Fine mapping of Leishmania major susceptibility Locus lmr2 and evidence of a role for Fli1 in disease and wound healing. Infect Immun. 2010;78(6):2734–2744. doi: 10.1128/IAI.00126-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salhi A, Rodrigues V, Jr, Santoro F, Dessein H, Romano A, Castellano LR, Sertorio M, Rafati S, Chevillard C, Prata A. Immunological and genetic evidence for a crucial role of IL-10 in cutaneous lesions in humans infected with Leishmania braziliensis. J Immunol. 2008;180:6139–6148. doi: 10.4049/jimmunol.180.9.6139. [DOI] [PubMed] [Google Scholar]
- Shar S, Lkhagvasuren D, Molur S. Rhombomys opimus. 2009. IUCN Red List of Threatened Species Version 2. http://www.iucnredlist.org.
- Smith A, Xie Y. The Mammals of China. Princeton University Press; Princeton, NJ: 2008. [Google Scholar]
- Sosnina EF. The lice of the gerbils of Tajikistan. Vshi peschanok Tadzhikistana. 1979;13:29–35. [PubMed] [Google Scholar]
- Strelkova MV, Eliseev LN, Ponirovsky EN, Dergacheva TI, Annacharyeva DK, Erokhin PI, Evans DA. Mixed leishmanial infections in Rhombomys opimus: A key to the persistence of Leishmania major from one transmission season to the next. Ann Trop Med Parasitol. 2001;95:811–819. doi: 10.1080/00034980120111154. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- WHO WHO: World Health Organisation internal report. 2009.
- WHO World Health Organization. 2012. < http://www.who.int/leishmaniasis/burden/en/. (accessed January 2012)
- Yaghoobi-Ershadi MR, Akhavan AA, Mohebali M. Meriones libycus and Rhombomys opimus (Rodentia: gerbillidae) are the main reservoir hosts in a new focus of zoonotic cutaneous leishmaniasis in Iran. Trans Roy Soc Trop Med Hyg. 1996;90:503–504. doi: 10.1016/s0035-9203(96)90295-3. [DOI] [PubMed] [Google Scholar]
- Yaghoobi-Ershadi MR, Javadian E. Epidemiological study of reservoir hosts in an endemic area of zoonotic cutaneous leishmaniasis in Iran. Bull World Health Organ. 1996;74:587–590. [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Dai X. Study on the situation of plague in Junggar Basin of China Study. Zhonghua Liu Xing Bing Xue Za Zhi. 2008;29:136–144. [PubMed] [Google Scholar]
- Ziaei H. A Field Guide for Identifying of Iranian Desert Mammalians. Iranian Environment Organization; Tehran: 1996. [Google Scholar]