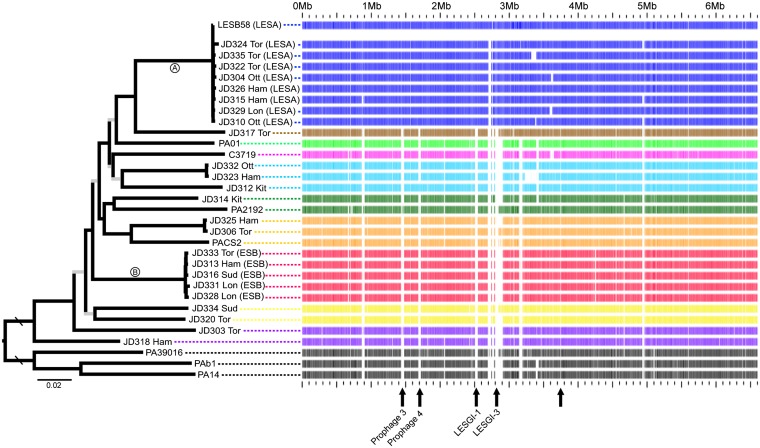
Fig. 2.
(Left) Maximum likelihood phylogeny constructed from the core genome alignment (5.06 Mb; 137,941 SNPs). All internal branches have bootstrap percentages >70 except those shown in gray. The rooting branch is four times longer than shown. Isolates with names starting with JD were sampled from Ontario (Ham, Hamilton; Kit, Kitchener; Lon, London; Ott, Ottawa; Sud, Sudbury; Tor, Toronto). (Right) Example of a BLAST atlas with a LESA isolate as the reference genome. Coding genes in the LESB58 genome are indicated along the Upper row as blue bars. For every other genome, the presence/absence of the corresponding LESB58 gene is indicated by presence/absence of a bar. Arrows along the bottom indicate regions of genome that were identified as LESA specific; named regions correspond roughly with regions identified in ref. 19.