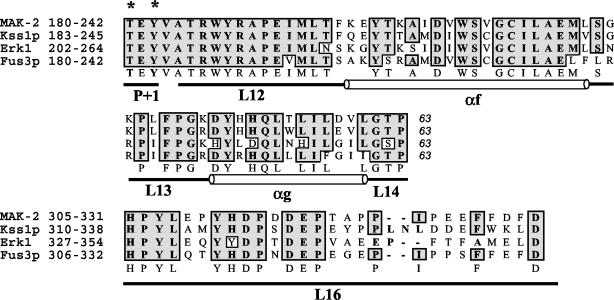
FIG. 1.
Alignment of MAK-2, Kss1p, Erk1, and Fus3p. All four of the MAP kinases are aligned by Clustal W with identity-based alignment. The regions presented are the catalytic core (63 amino acids, P+1 to L14) and the substrate interaction and docking domain (L16). All four of the MAP kinases are activated by phosphorylation at T*EY* in the P+1 loop in the catalytic domain. The phosphorylation lip starts from amino acids Asp-Phe-Gly (DFG) in subdomain VII and ends at TEY in subdomain VIII. Structural domains (L, loop; α, α-helix) are designated according established nomenclature (1). National Center for Biotechnology Information accession numbers are as follows: mak-2, AF348490; FUS3, Z35777; KSS1, Z72825; Erk1, P27361. Asterisks indicate phosphorylated residues.