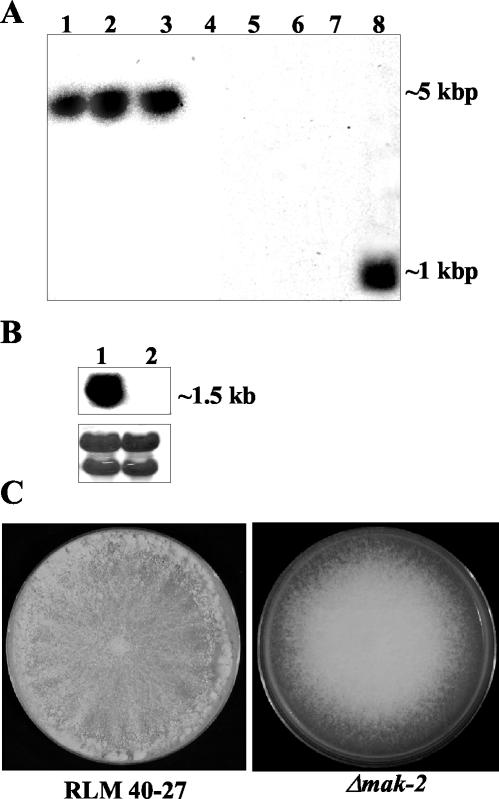
FIG. 2.
Analysis of wild-type and Δmak-2 strains. (A) Southern analysis of genomic DNA from Δmak-2 strains and RLM 40-27 (wild-type strain). Genomic DNA was digested with EcoRV and probed with a hygromycin phosphotransferase gene fragment (lanes 1 to 4) or a mak-2 PCR fragment (lanes 5 to 8). Δmak-2 progeny were APJ-2 (lanes 1 and 5), APJ-1 (lanes 2 and 6), PB-1 (lanes 3 and 7), and RLM 40-27 (lanes 4 and 8). (B) Northern analysis of mak-2 transcription in RLM 40-27 versus Δmak-2 strain PB-1. Two-day-old mycelia were used for RNA extraction. A mak-2 PCR fragment was used as a probe (see Materials and Methods). Lane 1, RLM 40-27; lane 2, PB-1. The same blot was stained with methylene blue as an RNA loading control (lower panel). The rRNA bands are shown. (C) Growth characteristics and morphology of a Δmak-2 mutant compared to wild-type (RLM 40-27). Conidia from RLM 40-27 and PB-1 were inoculated onto plates containing Vogel's (57) minimal medium and assessed for growth and conidiation. Δmak-2 mutants grow slower than the wild type (2.2 versus 7 cm/day) and have derepressed conidiation.