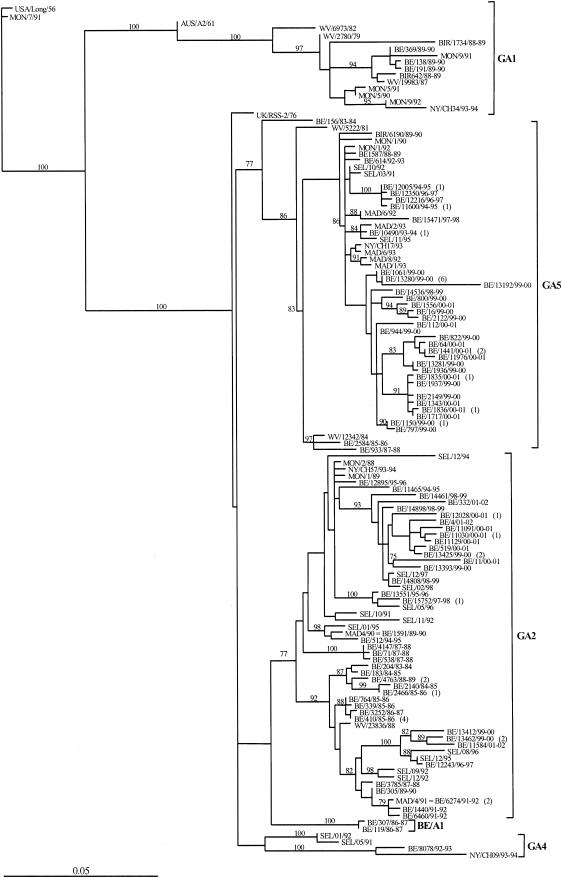
FIG. 1.
Phylogenetic tree of HRSV-A strains as computed with paup4b10. The nucleotide sequences of the G glycoprotein gene of the Belgian isolates were compared with those from Madrid (MAD), Montevideo (MON), Seoul (SEL), West Virginia (WV), and Birmingham (BIR) with the original nomenclature RSB89-642 (BIR642/88-89), RSB89-1734 (BIR1734/88-89), and RSB89-6190 (BIR6190/89-90) and compared with those from New York (NY) with original nomenclature CH17 (NY/CH17/93), CH09 (NY/CH09/93), CH57 (NY/CH57/94), CH34 (NY/CH34/94), and the reference strains Long (USA/Long/56), A2 (AUS/A2/61), and RSS-2 (UK/RSS-2/76). The Long strain was used as the outgroup sequence in the tree. The numbers at the internal nodes represent the number of bootstrap probabilities, as determined for 1,000 iterations by the neighbor-joining method. Only bootstrap values greater than 77% are shown. The italicized numbers in brackets at the terminal nodes correspond to the numbers of identical sequences. The genetic clusters obtained in the analysis are indicated by the square brackets and the designations GA1, GA2, GA4, GA5, and BE/A1.