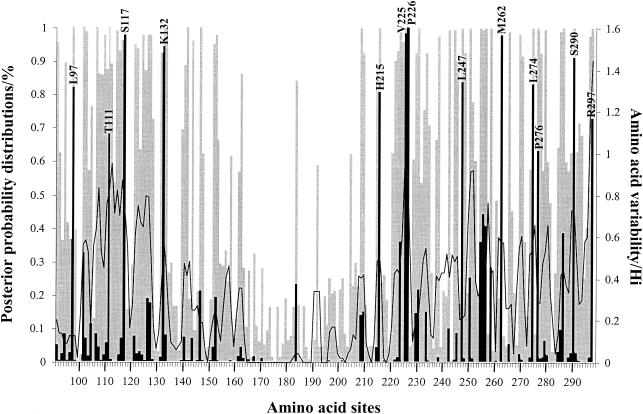
FIG. 2.
Posterior probabilities of site classes along the G protein ectodomain region under the discrete model M3. This model assumes three classes of sites in the gene: positive sites (▪), neutral sites (□), and negative sites ( ). Positively selected amino acid sites with posterior probabilities above 50% are indicated according to the Long strain. Amino acid variability, measured by entropy (Hi), is plotted to the second y axis.