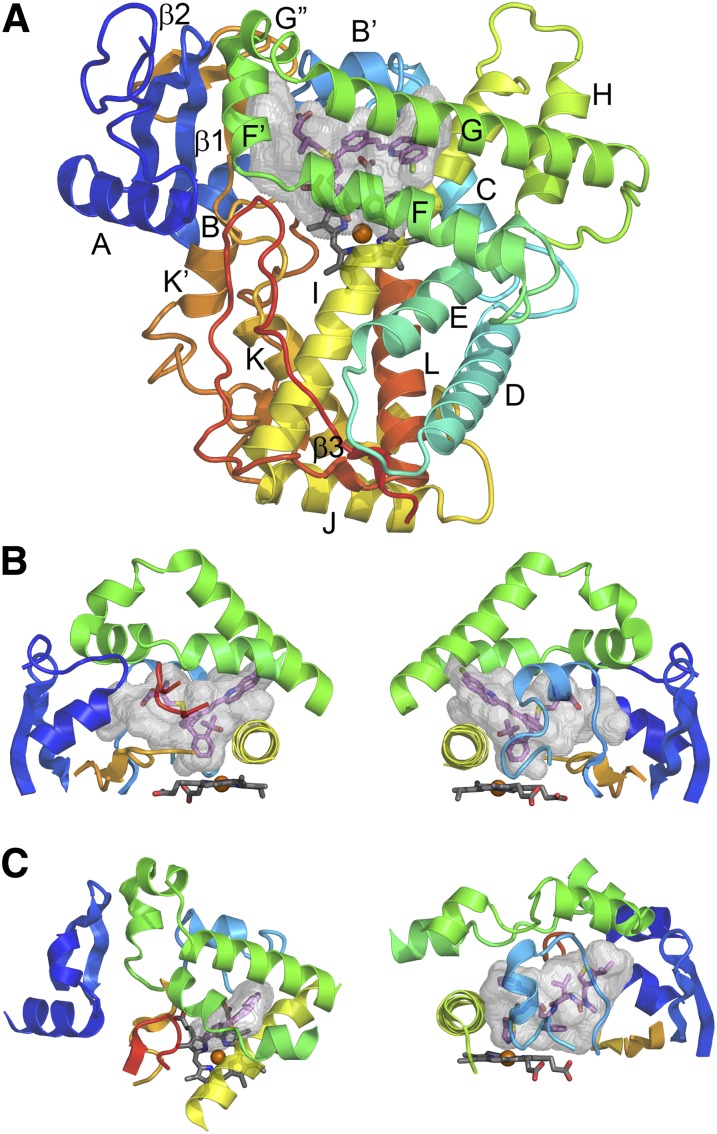
Fig. 3.
P450 secondary and tertiary structure. (A) Topological features of a microsomal P450 as illustrated by the Protein Data Bank:2NNI structure of human microsomal 2C8 colored from blue at the N-terminus to red at the C-terminus. The active site cavity is shown as a transparent surface. The bound substrate, montelukast (violet carbons), and the heme prosthetic group (gray carbons) are shown as stick figures. Twelve helices designated by letters A–L and β-sheets 1 and 2 are highly conserved. Additional helices are evident that are named by letters with prime or double prime designations. (B) Two views of structural components that form the sides of the substrate binding site of 2C8. The F–G region (green) forms the top of the cavity and is cantilevered over helix I (yellow) which forms one side. The opposite side is formed by connections (orange) between helix K and β1–3 and between β1–4 and helix K' near the surface of the heme, and by the N-terminal region (dark blue) that includes helix A and β-1. The gaps under the helix F–G region between helix I and the N-terminal region are filled by the C-terminal loop (red orange) as shown in the left panel and by the B–C loop (light blue) as shown in the right panel. (C) A view of the helix F–G side of human 1A2 α-napthoflavone complex, PDB:2HI4 (left) and of the B–C loop side of the human 3A4 ritonavir complex, Protein Data Bank:3NXU (right) illustrates differences in the topologies of the active sites and the secondary and tertiary structures of the three proteins. This figure was originally published in Johnson EF and Stout CD (2013) Structural diversity of eukaryotic membrane cytochrome P450s. J Biol Chem 288:17082–17090. ©The American Society for Biochemistry and Molecular Biology.