FIG. 4.
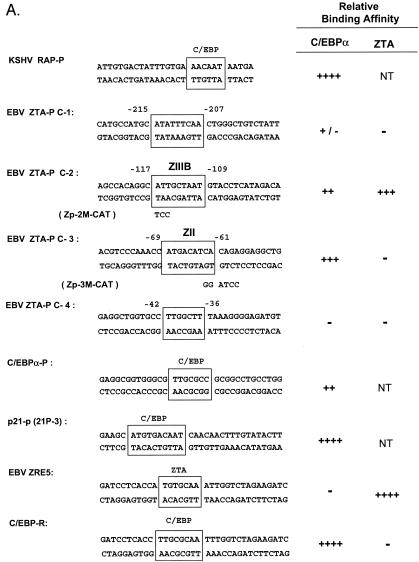
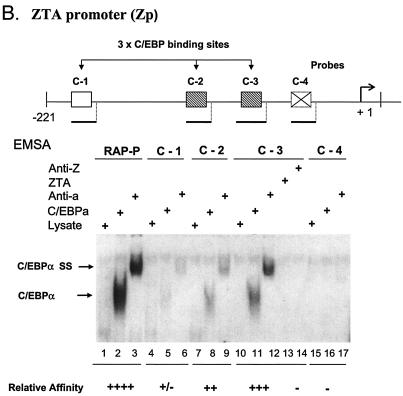
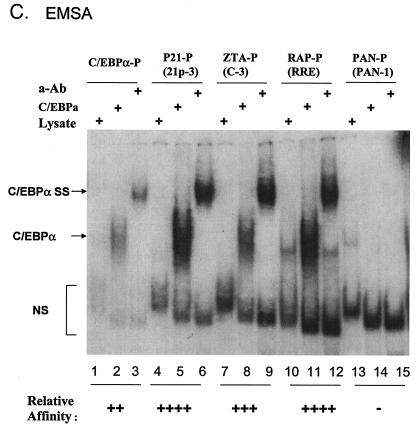
Identification of two strong C/EBP binding sites in the EBV Zp. (A) DNA sequences of double-stranded oligonucleotide probes used in EMSA experiments and semiquantitive summary of their relative affinities for C/EBPα: +, ++, +++, and ++++, approximate values of one, two-, four-, and eightfold, respectively; NT, not tested. Known or projected C/EBPα recognition motifs are boxed. Several probes containing C/EBP sites that were evaluated previously (8) or that were recently identified in various cellular or viral promoters are also included (71, 72). (B) EMSA experiment with Zp-derived oligonucleotide probes showing that in vitro-translated C/EBPα from pYNC172a binds more strongly to the C-3 site (lanes 11 and 12) than to the C-2 site (lanes 8 and 9), only weakly to the more distal C-1 site (lanes 5 and 6), and not at all to the C-4 site (lanes 16 and 17). Wild-type ZTA from pYNC100 also failed to bind to the C-3 site (lanes 13 and 14). The relative positions of the four C site probes between positions −221 and +1 in the proximal Zp are indicated in the upper diagram. Anti-Z, added anti-ZTA PAb; Anti-α, added anti-C/EBPα PAb; SS, supershift. (C) EMSA experiment comparing the binding affinities of the Zp C-3 site and selected oligonucleotide probes containing various other known C/EBP sites. C/EBPα-P (lanes 1 to 3) is the C/EBP binding site from the C/EBPα promoter; P21-P (21p-3; lanes 4 to 6) is the strongest C/EBP binding site from the p21CIP-1 promoter (71); ZTA-P (C-3; lanes 7 to 9) is the C/EBP C-3 site from the Zp; RAP-P (RRE; lanes 10 to 12) is the C/EBP-II site from the KSHV RAP promoter (66); and PAN-P (PAN-1; lanes 13 to 15) is a negative control sequence taken from the KSHV PAN promoter, which contains no C/EBP sites. In each group, the first lane contains an unprogrammed reticulocyte lysate, the second lane contains in vitro-translated C/EBPα (pYNC172a), and the third lane contains both C/EBPα and added anti-C/EBPα PAb. NS, nonspecific binding.