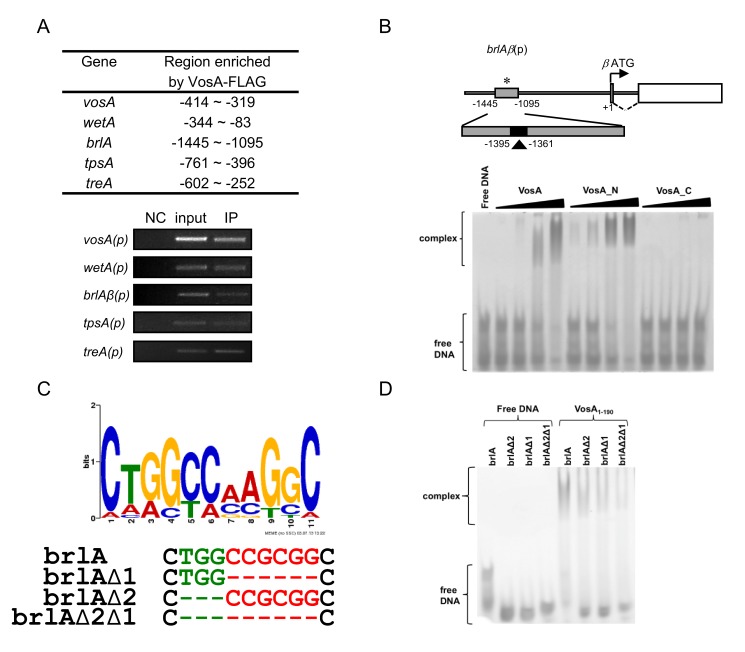
Figure 1. VosA binds DNA specifically.
(A) Selected regions enriched by VosA-FLAG ChIP on chip. The promoter regions of brlA, wetA, vosA, tpsA, and treA enriched by VosA-FLAG ChIP are shown (the start codon ATG is +1). Results of VosA-ChIP-PCR, the PCR amplicons separated on a 2% agarose gel, are shown in the bottom panel. The input DNA before immuno-precipitation (IP) was used as a positive control (input). The chromatin extract being incubated with bead only (without anti-FLAG antibody) was used as a negative control (NC). (B) Schematic presentation of the promoter region of brlA. The gray box (−1,095∼1,445 region of brlA(p), marked by *) represents the ChIP-PCR amplified region shown in (A), and the filled box (marked by arrowhead) represents a 35 bp fragment used in EMSA. EMSA using serially diluted VosA proteins and the 35 bp DNA probe of the brlA promoter (OHS301/302). DNA and protein were used in the molar ratios 1∶0.3, 1∶1, 1∶3, and 1∶9. VosA, full-length VosA; VosA_N, truncated VosA (residues 1–216); VosA_C, truncated VosA (residues 217–430). (C) The consensus DNA sequence predicted to be recognized by VosA is shown. Two cores of this motif were deleted as shown (green, red) and these probes were used for additional EMSA. (D) EMSA using the crystallized VosA1–190 (VosA_cryst) with wild-type and mutated DNA probes of the brlA promoter. In the mutated versions of the DNA, the predicted VosA binding motifs 1 and 2 were deleted (brlA, wild-type DNA (OHS301/302); brlAΔ2, DNA with deleted core 2 (JG636/637); brlAΔ1, DNA with deleted core 1 (JG638/639); brlAΔ2Δ1, DNA with deleted cores 2 and 1 (JG640/641)).