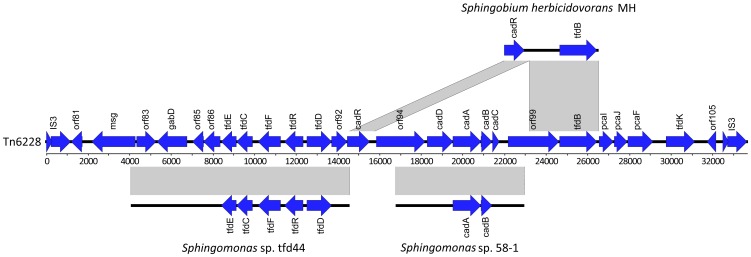
Figure 2. Genetic organization of the composite transposon Tn6228 and comparison to other gene clusters.
Genes are marked with blue arrows. Gene abbreviations are msg: malate synthase G, gabD: putative succinate-semialdehyde dehydrogenase, tfdE: dienelacatone hydrolase, tfdC: chlorocatechol 1,2-dioxygenase, tfdF: maleylacetate reductase, tfdR: LysR family transcriptional regulator, tfdD: chloromuconate cycloisomerase, tauE/safE: sulfite exporter, cadR: AraC family transcriptional regulator, cadD: oxidoreductase component of Rieske non-heme iron oxygenase (RO), cadA: large subunit of 2,4-D oxygenase, cadB: small subunit of 2,4-D oxygenase, cadC: ferredoxin component of RO, tfdB: dichlorophenol hydroxylase, pcaI: 3-oxoacid CoA-transferase A subunit, pcaJ: 3-oxoacid CoA-transferase B subunit, pcaF: beta-ketoadipyl CoA thiolase, tfdK: transport protein. Nucleotide similarity is shown as gray bars linking to the corresponding gene clusters in Sphingomonas sp. tfd44 (acc. no. AY598949.1), Sphingomonas sp. 58-1 (acc. no. AB353895.1) and Sphingobium herbicidovorans MH (acc. no. AJ628861.1). Nucleotide similarities are shown with a cutoff value of 98% and were identified with BLASTN as implemented in Easyfig [27]. The composite figure shown here was compiled in Inkscape [28].