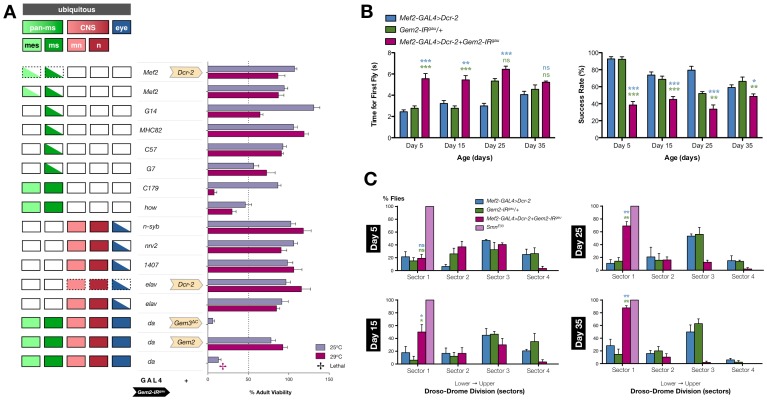
Figure 8. Gemin2 is essential for viability and normal motor function.
(A) Gemin2 knockdown through the expression of a single RNAi transgene (Gem2-IRgau) causes a drastic reduction in adult viability (25°C) or complete lethality (29°C) if driven in all tissues via the da-GAL4 driver. Expression of transgenic Gemin2 within this genetic background restores viability. No significant rescue is however observed on co-expression of an unrelated transgene (Gem3 ΔC). A marked reduction in adult viability can also be observed if Gemin2 is depleted specifically within muscle lineages (C179-GAL4 or how-GAL4). Note that, as expected, not all drivers with a similar tissue expression pattern (left panel) give an analogous result with one reason being that they might not drive strong GAL4 levels comparable to their effective counterparts. Bar chart (right panel) shows adult fly viability assayed at 25°C and 29°C, with the latter temperature inducing maximal GAL4 activity. Abbreviations: pan-ms, pan-muscular; mes, mesoderm; ms, larval muscles; mn, motor neurons; n, all CNS neurons except motor neurons. (B) Pan-muscular Gemin2 knockdown boosted by enhanced Dicer-2 levels leads to impaired climbing behaviour. In this respect, the time taken for the first fly within the sampled population to reach a target height of 8 cm is significantly increased on both day 5 and day 15 post-eclosion. In a similar vein, the climbing success rate or the percentage of flies that reach beyond 8 cm within 10 seconds, was dramatically decreased at all the recorded time points. (C) Flies with muscle-specific Gemin2 depletion exhibit a progressive age-dependent decline in flight performance. In this regard, the portion of flightless flies is significantly different from controls on day 15 post-eclosion and increases with age until reaching a maximal level on day 35 of adulthood. On the latter time point, the behaviour is similar to that of flies with reduced levels of SMN in muscle (SmnE33), which are 100% non-fliers. For all data, individual bars represent the mean ± S.E.M. and statistical significance was determined for differences between the Mef2-GAL4>Dcr-2+Gem2-IRgau genotype and control genotypes, which are indicated by the respective colour (B, C). For all data, ns = not significant, *p<0.05, **p<0.01, and ***p<0.001.