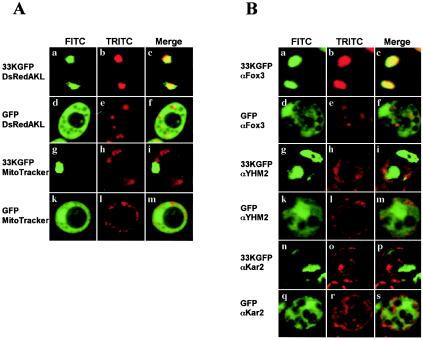
FIG. 1.
Fusion protein p33GFP localizes to peroxisomes in S. cerevisiae. (A) Yeast cells were transformed with a plasmid expressing fused p33GFP (a to c and g to i) or free GFP (d to f and k to m). The cells in panels a to c and d to f were cotransformed with a plasmid expressing the protein DsRedAKL localizing on peroxisomes. The cells in panels g to i and k to m were treated with the mitochondrial stain MitoTracker. Living cells were analyzed by fluorescence microscopy, and representative images for the FITC (left column, green), TRITC (middle column, red) channels, and merged signals (right column) are shown. (B) Cells of the same transformants as in panel A expressing fused p33GFP (a to c, g to i, and n to p) or free GFP (d to f, k to m, and q to s) were fixed and immunostained with antibodies to proteins localizing to peroxisomes (Fox3), mitochondria (YHM2), or the ER (Kar2). Representative images for GFP (left column, FITC), rhodamine (middle column, TRITC), and merged signals (right column) are shown.