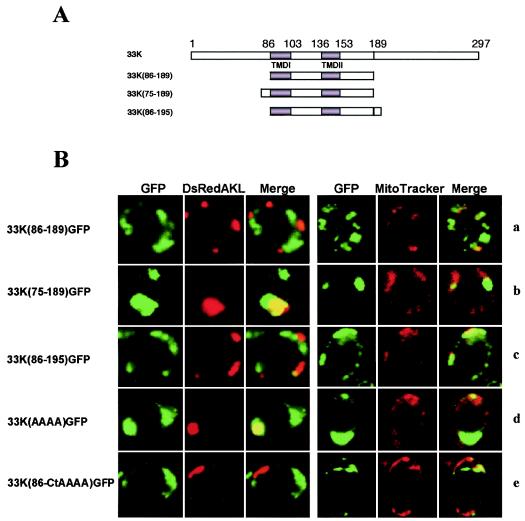
FIG. 6.
Expression of GFP in yeast cells transformed with different mutants of the p33 gene obtained by deletions in the N- and C-terminal hydrophilic regions. (A) Diagram of each mutant with the indication of the positions of the TMDs and the relevant restriction site SacI; (B) selected images of each mutant for GFP, DsRed targeted to peroxisomes or mitochondria (DsRedAKL and MitoTracker, respectively), and merged signals. In addition, images for clones 33K(AAAA)GFP and 33K(86-CtAAAA)GFP are shown; these are the same as 33KGFP and 33K(86-Ct)GFP, respectively, except that amino acids RRRR at positions 213 to 216 were mutated to AAAA. The clone 33K(75-189)GFP (row b) expresses the shortest protein containing the complete signal for peroxisomal targeting. Conversely, proteins expressed from clones 33K(86-189) and 33K(86-195) (rows a and c, respectively) are essentially cytoplasmic, although protein p33(86-195) shows a weak mitochondrial targeting. Peroxisomal targeting of the clone 33K(86-Ct)GFP (see Fig. 5) was abolished if the tetrapeptide RRRR was mutated to AAAA [clone 33K(86-CtAAAA)GFP] (row e). These same amino acids are irrelevant in the full-length context of p33 [clone 33K(AAAA)GFP] (row d).