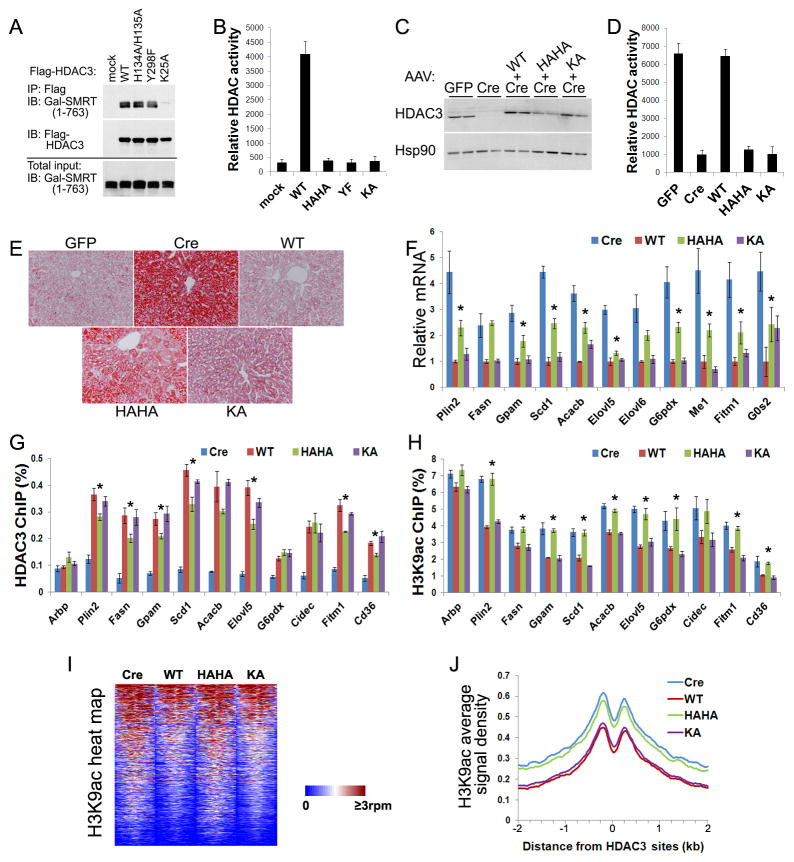
Figure 4. Deacetylase-dead HDAC3 rescues HDAC3-dependent transcriptional repression despite failing to repress genome-wide histone acetylation.
(A) GAL-tagged SMRT (1–763) and Flag-HDAC3 were co-expressed in HEK 293T cells. Cell lysates were immunoprecipitated with Flag antibodies followed by immunoblot analysis. (B) HDAC assay of the above immunoprecipitates in triplicates. (C) HDAC3f/f mice were injected with AAV-Tbg-GFP or AAV-Tbg-Cre along with AAV vectors expressing Flag-tagged HDAC3 mutants. Total liver lysates were analyzed by immunoblot analysis. (D) Liver lysates were immunoprecipitated with anti-HDAC3 antibodies and assayed for enzyme activities, n = 4. (E) ORO staining of livers. (F) RT-qPCR analysis of livers, n = = 4. * P < 0.05 between Cre and HAHA. (G) Livers were subjected to chromatin immunoprecipitation (ChIP) with HDAC3 antibodies followed by qPCR analysis using primers for HDAC3 sites near the indicated genes, n = 4. * P < 0.05 between WT and HAHA. (H) ChIP-qPCR analysis with antibodies against acetylated lysine 9 of histone 3 (H3K9ac), n = 4. * P < 0.05 between WT and HAHA. (I) Heat map of H3K9ac ChIP-seq signals from −1 kb to +1 kb surrounding the center of HDAC3 sites within 50 kb of genes that are upregulated upon HDAC3 depletion. rpm, reads per million. (J) Average H3K9ac signals, represented by reads per bp in 10 million total reads, from −2 kb to +2 kb surrounding the center of the same HDAC3 sites as shown above. G0S2, G0/G1 switch 2; Arbp, ribosomal protein, large, P0, also known as 36B4. All error bars, s.e.m. See also Figure S5.