Table 1. Quality measures for the reconstructed hierarchies in case of the protein function data set.
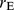
|
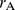
|
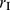
|
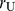
|
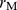
|
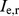
|
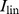
|
|
| algorithm A | 21% | 66% | 2% | 32% | 0% | 35% | 78% |
| algorithm B | 20% | 52% | 3% | 44% | 1% | 30% | 75% |
| P. Heymann & H. Garcia-Molina | 19% | 51% | 3% | 46% | 0% | 30% | 75% |
| P. Schmitz | 18% | 65% | 2% | 23% | 10% | 30% | 75% |
The quality of the tag hierarchy obtained for the tagged protein data set, 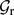 , was evaluated by comparing it to the hierarchy of protein functions in the Gene Ontology,
, was evaluated by comparing it to the hierarchy of protein functions in the Gene Ontology, 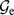 . The quality measures presented in the different columns are the following: the ratio of exactly matching links in
. The quality measures presented in the different columns are the following: the ratio of exactly matching links in 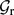 ,denoted by
,denoted by 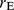 , the ratio of acceptable links,
, the ratio of acceptable links, 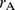 , (connecting more distant ancestor-descendant pairs), the ratio of inverted links,
, (connecting more distant ancestor-descendant pairs), the ratio of inverted links, 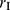 , (pointing in the opposite direction), the ratio of unrelated links,
, (pointing in the opposite direction), the ratio of unrelated links, 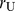 , (connecting tags on different branches in
, (connecting tags on different branches in 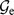 ), the ratio of missing links in
), the ratio of missing links in 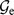 , denoted by
, denoted by 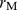 , the normalized mutual information between the two hierarchies,
, the normalized mutual information between the two hierarchies, 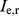 , and the linearized mutual information,
, and the linearized mutual information, 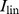 , corresponding to the fraction of exactly matching links remaining after a random link rewiring process stopped at NMI value given by
, corresponding to the fraction of exactly matching links remaining after a random link rewiring process stopped at NMI value given by 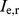 . The different rows correspond to results obtained from algorithm A (1
. The different rows correspond to results obtained from algorithm A (1 row), algorithm B (2
row), algorithm B (2 row),the method by P. Heymann & H. Garcia-Molina (3
row),the method by P. Heymann & H. Garcia-Molina (3 row), and the algorithm by P. Schmitz (4
row), and the algorithm by P. Schmitz (4 row).
row).
