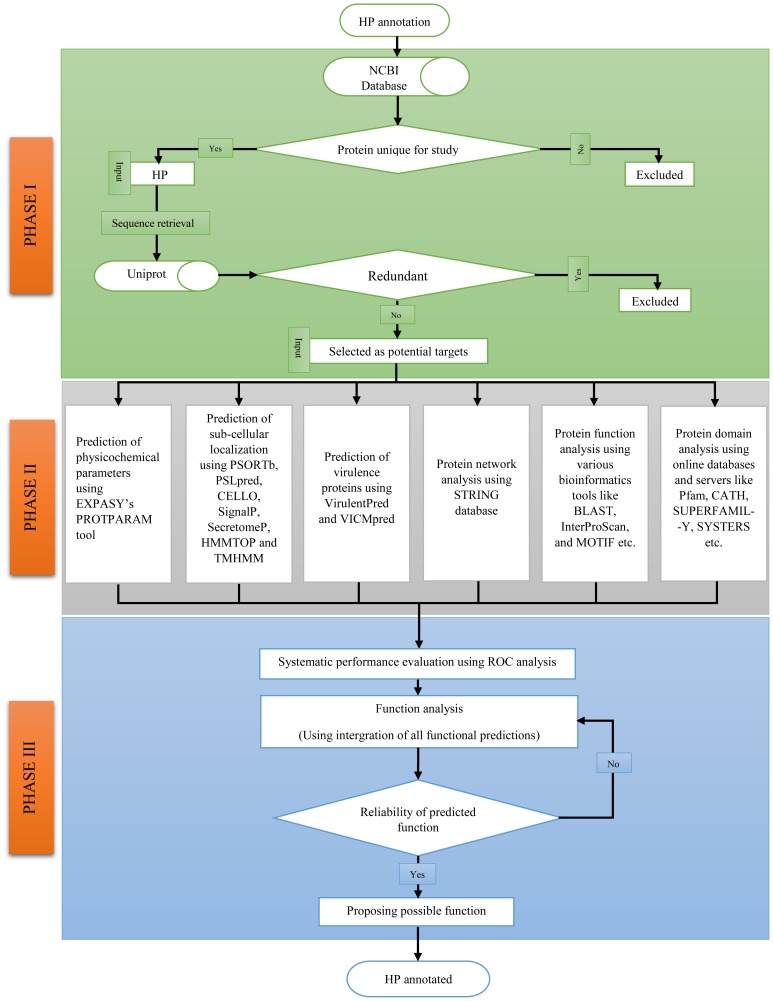
Figure 1. Computational framework used for annotating function of 429 HPs from H. influenzae.
Methodology is divided into three phases: PHASE I. H. influenzae HP characterization and sequence retrieval from online databases. PHASE II. The extensive analysis of sub-cellular localization, physicochemical parameters, virulence, function and domain present in HPs. PHASE III. This phase include assessment of predicted functions using the protein with known function from H. influenzae and reliable prediction of possible functions of HPs.