Abstract
Traumatic brain injury (TBI) is an increasingly important public health concern. While there are several promising avenues of intervention, clinical assessments are relatively coarse and comparative quantitative analysis is an emerging field. Imaging data provide potentially useful information for evaluating TBI across functional, structural, and microstructural phenotypes. Integration and management of disparate data types are major obstacles. In a multi-institution collaboration, we are collecting electroencephalogy (EEG), structural MRI, diffusion tensor MRI (DTI), and single photon emission computed tomography (SPECT) from a large cohort of US Army service members exposed to mild or moderate TBI who are undergoing experimental treatment. We have constructed a robust informatics backbone for this project centered on the DICOM standard and eXtensible Neuroimaging Archive Toolkit (XNAT) server. Herein, we discuss (1) optimization of data transmission, validation and storage, (2) quality assurance and workflow management, and (3) integration of high performance computing with research software.
Keywords: traumatic brain injury, automatic multi-modal image analysis, XNAT
1. INTRODUCTION
Traumatic brain injury (TBI), characterized by altered mental status (e.g., short-term memory and concentration difficulties), is an important medical concern for combat-zone military personnel, especially for soldiers with blast injuries [1]. In addition, TBI is the most common cause of death and disability in young people [2]. Although some progress in diagnosis and treatment of TBI patients has been achieved, clinical evaluation is relatively coarse and reliable quantitative assessments of mild to moderate injury have yet to be developed. Modern imaging techniques may contribute to characterizing TBI by providing structural, functional and metabolic information. In our project, quantitative electroencephalography (EEG), T1-weighted MRI (T1w), diffusion weighted MRI (DWI), single photon emission computed tomography (SPECT) and behavioral test data are collected from a large cohort of US Army service members exposed to mild or moderate TBI who are undergoing experimental treatment. However, no fully customized system exists to transmit, manage and analyze our large volume of data from different sources.
This paper presents the framework of an integrated system customized for our TBI project. The system is centered on the DICOM standard and eXtensible Neuroimaging Archive Toolkit (XNAT) [3] server to facilitate multi-modal data storage and access from multiple institutions. The system uses a cluster environment to perform distributed computing for high throughput data analysis. We discuss the overall system structure and the key design elements including (1) optimization of data transmission, validation and storage, (2) quality assurance and workflow management, and (3) integration of high performance computing with diverse research software packages.
2. METHODS AND RESULTS
2.1 System Overview
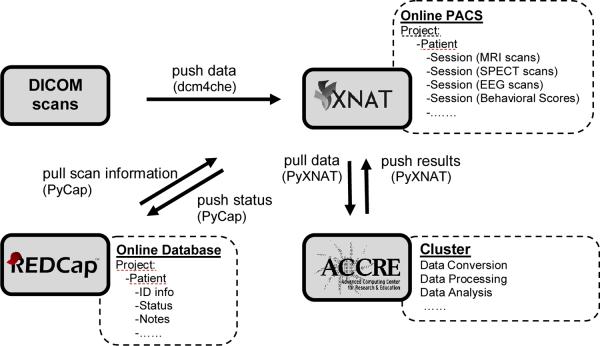
The proposed system integrates three main server resources designed for different functions (Figure 1): 1) An XNAT/PACS server supporting the transfer and storage of image data, especially DICOM data; 2) A Research Electronic Data Capture (REDCap) server storing all the related demographic data; 3) the Advanced Computing Center for Research and Education (ACCRE) server providing a cluster environment for high performance distributed computing. This effort re-envisions and extends our earlier work on PACS-cluster integration [4]. Data communication among these servers is implemented primarily by a set of scheduled tasks implemented in the Python programming language running on a CentOS 64-bit coordinating server. The original DICOM images, acquired from multi-modality scanners, are routed to XNAT/PACS via open source DCM4CHE tools [5]. Then the images stored on XNAT are retrieved via python scripts using the PyXNAT package (http://packages.python.org/pyxnat/) and processed on the cluster using several standard and custom research software packages. The processing results are pushed back to XNAT. Scan information is pulled from and pushed to the REDCap database via the PyCap package (http://pypi.python.org/pypi/PyCap/).
Figure 1.
Overview of the integrated system. Multi-modal DICOM scans (upper left) are pushed to the XNAT/PACS server (upper right) via dcm4che tools. Verified DICOM scans are pulled and distributed to the cluster (bottom right) for further conversion, processing, and analysis by PyXNAT package scripts. The output results are pushed back for storage on XNAT. The online database REDCap (bottom left) provides/stores related information to/from XNAT. Black arrows indicate the directions of the data flow.
2.2 Optimization of data transmission, validation and storage
The original DICOM files are transmitted from distributed clients to a central XNAT/PACS server using the DCM4CHE library, which is able to send all DICOM files automatically under a specified directory. The XNAT/DICOM gateway routes every uploaded DICOM file to the correct storage folder according to DICOM header information (i.e. Patient ID, Modality, Study UID and Series UID, etc.). The XNAT/DICOM gateway also serves as a query/retrieve service class provider (SCP) supporting transmission of DICOM files and other formats of data from the XNAT server to the client.
Uploaded DICOM datasets could be corrupted on XNAT/PACS if any transmission errors occur, thus the integrity of the dataset needs to be verified before any further operations are performed on the data. The supposed number of DICOM files in each original scan is able to be acquired from DICOM file header (Tag-<0020, 1002>, Attribute Name-<images in acquisition>). This number is compared to the actual number of DICOM files stored on XNAT. A discrepancy stops further processing on the scan until manually corrected. The validation protocol is automatically executed by a scheduled task running on the coordinating server.
The file storage system of XNAT has top-to-bottom levels: project, subject, experiment, scan and resource. All our data are stored under a TBI project. A subject folder is created for each patient with a unique subject ID. An experiment holds the data acquired and processed from each experimental modality (e.g., MRI, SPECT and EEG) for one patient. Scans with specific acquisition settings (e.g., T1w and DWI scans) from the same modality all go into the same experiment. The results computed from scan data by a specified processing protocol are stored as a new scan in the same experiment as the original scan. For those scans holding original verified DICOM data as a resource, the data converted to another format (e.g., NIfTI) is stored as a parallel resource in the same scan to accommodate input requirements of specific software routines.
2.3 Quality assurance and workflow management
Large volumes of imaging data require the automation of quality assurance (QA). Currently, three primary protocols for T1w anatomy QA, DTI QA and registration QA are running in the system. The T1w anatomy QA displays the parcellation results computed from FreeSurfer software [6, 7] (http://surfer.nmr.mgh.harvard.edu/), as well as measurements of reliability including the contrast-to-noise ratio (CNR) across the gray/white matter boundary surface by region and agreement between the white matter surface and gray matter segmentation. The DTI QA protocol mainly consists of automatic estimation of head motion and Fractional Anisotropy (FA) variance and bias [8] by statistical methods [9], assessing quality via power calculations and normative values, and robust fractional anisotropy calculation (see more details in [10]). The registration QA protocol includes automatic estimating the quality of skull striping and display the final registration results in blend mode.
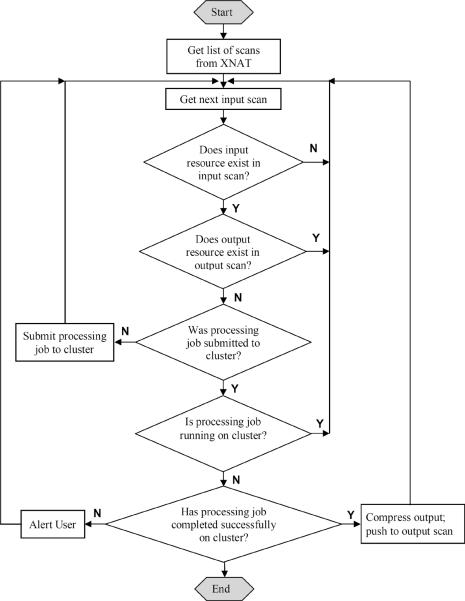
Workflow is automatically monitored by a set of scheduled tasks in Python running on the coordinating server. Those periodically activated tasks are responsible for checking the status of each input scan on XNAT and then triggering corresponding actions. Basically, one verified input scan will pass through three processing states as the job is 1) not yet started; 2) running; and 3) completed. The typical logic of the management task is shown in Figure 3.
Figure 3.
Flowchart of distributed job management. (Note the `job' in the flowchart refers to one of the jobs listed in Table 1; the `input' and `output' refer to arguments and output results of the processing job, respectively).
2.4 Integration of high performance computing with research software
To increase the efficiency of system development and computing performance, a number of mature applications are wrapped into processing jobs which are executed on the CentOS 6 compute nodes of the ACCRE cluster. All processing steps are designed for the final goal of identifying correlations among imaging and behavioral data, to evaluate new approaches to both diagnosis and detection of treatment effects.
Table 1 lists the function, input, output, wrapped software and average running time of current and prospective processing jobs. One function of job group 1 is to extract anatomical information from the high resolution `T1w scan' (1×1×1mm3 voxel size) via FreeSurfer v5 [6, 7, 11–14] (FS, http://surfer.nmr.mgh.harvard.edu/). The `FS data' is a ZIP file of compressed FS output including cortical labels, pial/white matter surface meshes and so on. This is stored as a new `scan' on XNAT. The other function of job group 1 is quality assurance of the anatomical measurements calculated by FS. The anatomical QA is implemented in MATLAB 2012a (Mathworks, Natick, MA). The output `anatQA data' includes a PDF QA report shown in Figure 2A and MAT files of QA variables. The purpose of job group 2 is to align the reference T1w image space of each subject to a target DWI space (acquired by a spin echo EPI sequence, 1×1×2mm3 voxel size) of the same subject and to the MNI152 common space (1×1×1mm3 voxel size). The intra-subject affine registration with 12 degrees of freedom is implemented by FMRIB's Linear Image Registration Tool v5.5 [15, 16] (FLIRT, http://fmrib.ox.ac.uk/fsl/flirt/) and the inter-subject non-linear registration is completed via Symmetric Normalization (SyN) [17] in the Advanced Normalization Tools v1.00 (ANTs, http://picsl.upenn.edu/ANTS/). The skull stripping prior to the registration is carried out by Brain Extraction Tool v2.1 [20] (BET, http://fmrib.ox.ac.uk/fsl/bet2/). Output results include transformed images, the deformation matrix and registration QA report shown in Figure 2C. Job Group 4 has the same design as group 2 except that the reference space is a SPECT volume with 2.5×2.5×2.5mm3 voxel size rather than the T1w space. Job group 3 is composed of three DTI related jobs: 1) quality assurance written in MATLAB wrapping Camino Diffusion MRI Toolkit v1038 [18, 19] (Camino, http://cmic.cs.ucl.ac.uk/camino/) and BET; 2) probabilistic tractography to obtain connectivity information via FMRIB's Diffusion Toolbox v2.0 [21] (FDT, http://fmrib.ox.ac.uk/fsl/fdt/); 3) tract-based spatial statistics (TBSS) analysis using TBSS toolkit v1.2 [22, 23] (http://fmrib.ox.ac.uk/fsl/tbss/). Job group 5 will conduct multimodal analyses across all types of imaging data and behavioral test scores. The output `scans' on XNAT will store all correlation results.
Table 1.
List of processing jobs and their corresponding function, input/output and wrapped research software.
| Job Group | Job Function | Job Input | Job Output | Wrapped software | Time |
|---|---|---|---|---|---|
|
| |||||
| 1* | Anatomical Analysis | T1w scan | FS data | FreeSurfer | ~1day |
| Anatomical Quality Assurance | FS data | anatQA data | MATLAB | ~15min | |
| 2* | Intra-subject Registration & Quality Assurance | T1w scan; DWI scan | T1toDWI data | BET, FLIRT | ~5min |
| Inter-subject Registration & Quality Assurance | T1w scan; MNI atlas | T1toMNI data | BET, ANTS (SyN) | ~5min | |
| 3* | DTI Analysis & Quality Assurance | DWI scan | dtiQA data | Camino, BET, MATLAB | ~1day |
| DTI Tractography | DWI scan; FS data; T1toDWI data | FDT data | FDT | ~2day | |
| Tract-Based Spatial Statistics | dtiQA data | TBSS data | TBSS | ~3day | |
| 4** | Intra-subject Registration & Quality Assurance | SPECT scan; DWI scan | SPECTtoDWI data | BET, FLIRT | - |
| Inter-subject Registration & Quality Assurance | SPECT scan; MNI atlas | SPECTtoMNI data | ANTS (SyN) | - | |
| 5** | Correlate MRI with SPECT/EEG/Behavioral Scores | All the output data | Correlation data | MATLAB | - |
current processing jobs
prospective processing jobs
Figure 2.
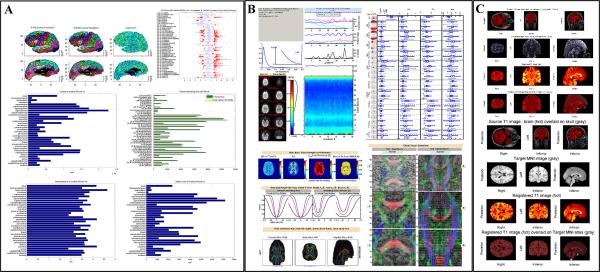
Example of quality assurance (QA) reports: (A) T1w Anatomy QA for the left hemisphere, (B) DTI QA for one DTI scan and (C) registration QA for aligning T1w space to DWI space as well as to MNI common space.
3. CONCLUSION AND DISCUSSION
Our system centers on XNAT/PACS with an optimized plan for data transmission, validation and storage. The system also integrates multiple mature/self-developed applications with high performance computation and implements quality assurance and automatic workflow to accommodate a large volume of multi-modal image data.
Although the system aims to automate the entire workflow, unexpected interruptions inevitably occur due to limitations of the hardware and research software configurations. Thus, manual intervention is needed to further analyze and correct problems identified by the system (Figure 3). For system interruptions (e.g. internet disconnection, server dysfunction and system resources deficiency), the affected jobs will be automatically resubmitted after system recovery. For research software errors, case-by-case studies are needed. In particular, image characteristics vary over a range, so some software parameters (e.g. thresholds) should be carefully selected so the software is able to deal with as many datasets as possible.
ACKNOWLEDGEMENTS
This project was supported by contract #W91YTV-12-C-0015 from the U.S. Army. The authors thank Alexander Dagley, Andrew Asman and Michael Esparaza for their assistance. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. This work described herein has not been submitted elsewhere for publication or presentation.
Footnotes
Publisher's Disclaimer: This publication has been reviewed and approved by BACH Public Affairs Officer and Operations Security Manager for compliance with operational security (OPSEC) guidance and regulations. The information in this publication does not constitute official endorsement by the U.S. Army or the Army Medical Command.
REFERENCES
- [1].Hoge CW, McGurk D, Thomas JL, et al. Mild traumatic brain injury in US Soldiers returning from Iraq. New England Journal of Medicine. 2008;358(5):453–463. doi: 10.1056/NEJMoa072972. [DOI] [PubMed] [Google Scholar]
- [2].Ghajar J. Traumatic brain injury. Lancet. 2000;356(9233):923–929. doi: 10.1016/S0140-6736(00)02689-1. [DOI] [PubMed] [Google Scholar]
- [3].Marcus DS, Olsen TR, Ramaratnam M, et al. The Extensible Neuroimaging Archive Toolkit - An informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5(1):11–33. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- [4].Esparza ML, Welch EB, Landman BA. Automating PACS quality control with the Vanderbilt image processing enterprise resource. 8319:83190H–7. doi: 10.1117/12.910800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Warnock MJ, Toland C, Evans D, et al. Benefits of using the DCM4CHE DICOM archive. Journal of Digital Imaging. 2007;20:125–129. doi: 10.1007/s10278-007-9064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- [7].Fischl B, van der Kouwe A, Destrieux C, et al. Automatically parcellating the human cerebral cortex. Cerebral Cortex. 2004;14(1):11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- [8].Lauzon CB, Crainiceanu C, Caffo BC, et al. Assessment of bias in experimentally measured diffusion tensor imaging parameters using SIMEX. Magnetic Resonance Medicine. 2012 doi: 10.1002/mrm.24324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Whitcher B, Tuch DS, Wisco JJ, et al. Using the wild bootstrap to quantify uncertainty in diffusion tensor imaging. Human Brain Mapping. 2008;29(3):346–362. doi: 10.1002/hbm.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lauzon CB, Caffo BC, Landman BA. Towards automatic quantitative quality control for MRI. 8314:83140K–8. doi: 10.1117/12.910819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis - I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- [12].Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis - II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- [13].Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- [14].Fischl B, Salat DH, van der Kouwe AJW, et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage. 2004;23:S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- [15].Jenkinson M, Bannister P, Brady M, et al. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. NeuroImage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- [16].Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Medical Image Analysis. 2001;5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- [17].Avants BB, Epstein CL, Grossman M, et al. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cook PA, Bai Y, Nedjati-Gilani S, et al. Camino: Open-Source Diffusion-MRI Reconstruction and Processing. 2759 [Google Scholar]
- [19].Chang L-C, Jones DK, Pierpaoli C. RESTORE: Robust estimation of tensors by outlier rejection. Magnetic Resonance in Medicine. 2005;53(5):1088–1095. doi: 10.1002/mrm.20426. [DOI] [PubMed] [Google Scholar]
- [20].Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Behrens TEJ, Woolrich MW, Jenkinson M, et al. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magnetic Resonance in Medicine. 2003;50(5):1077–1088. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- [22].Smith SM, Jenkinson M, Johansen-Berg H, et al. Tract-based spatial statistics: Voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31(4):1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- [23].Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23(Supplement 1(0)):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]