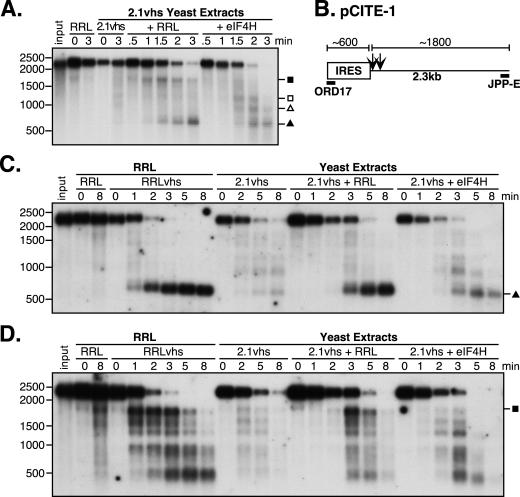
FIG. 7.
eIF4H does not restore efficient IRES-directed targeting to yeast-expressed vhs. (A) Analysis of nuclease activity on pCITE-1 RNA. Internally labeled pCITE-1 RNA was added to control RRL (RRL) and yeast extracts containing 2.1vhs in the presence or absence of RRL or eIF4H (2.1vhs, 2.1vhs+RRL, and 2.1vhs+eIF4H). Samples were processed as described in the legend for Fig. 1. (B) Diagram of pCITE-1 RNA, indicating the positions of hybridization of oligonucleotides ORD17 and JPP-E specific for the 5′ and 3′ ends of the RNA, respectively. Also indicated are the RNA fragments produced upon initial cleavage of the RNA by vhs translated in RRL. (C and D) Northern blot analysis of nuclease activity on pCITE-1 RNA using oligonucleotide probes for the 5′ and 3′ ends, respectively, of pCITE-1 RNA. Unlabeled pCITE-1 RNA was added to control RRL (RRL), RRL containing pretranslated vhs (RRLvhs), and yeast extracts containing 2.1vhs, either alone (2.1vhs), mixed with RRL (2.1vhs+RRL), or mixed with eIF4H (2.1vhs+eIF4H). RNA was extracted at the indicated time points, resolved on an agarose-formaldehyde gel, transferred to a Gene Screen Plus membrane, and analyzed by Northern blot analysis. Symbols are as described in the legend for Fig. 3. The closed square indicates the position of the 1,800-nt fragment. The positions and sizes of the RNA markers in nucleotides are at the left.