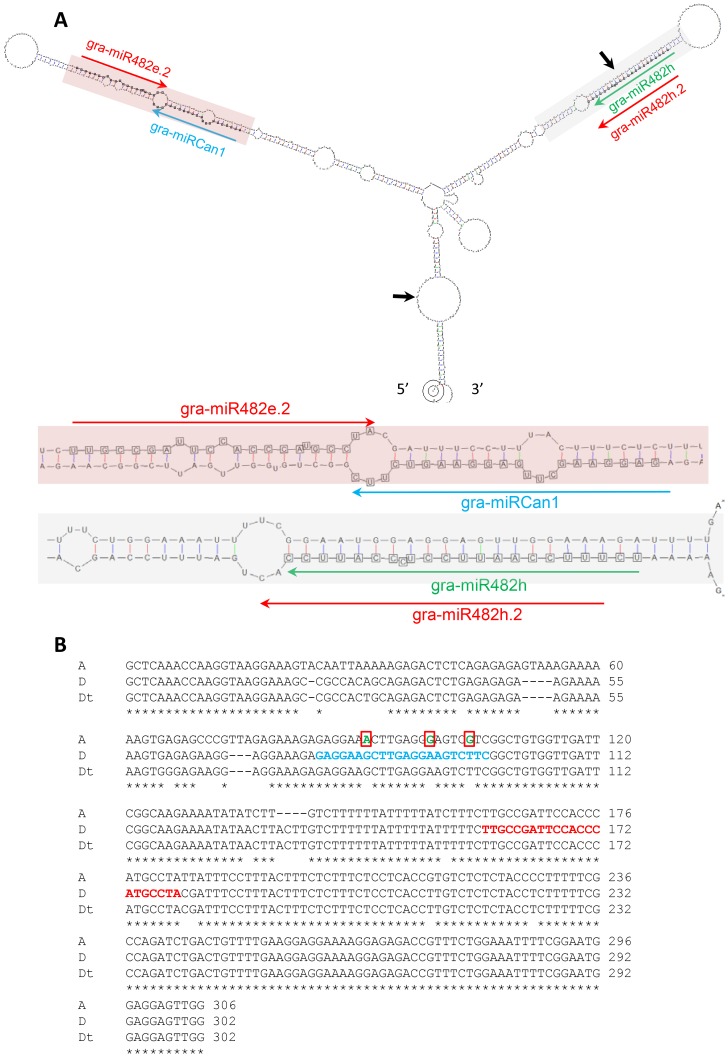
Figure 2. A single primary MIRNA transcript contains gra-miR482h/miR482h.2 and gra-miR482e.2.
A) Hairpin structure of the precursor (G. raimondii) containing gra-miR482h/miR482h.2 and gra-miR482e.2. The details of the shaded regions of the precursor are shown below the hairpin structure. B) Aligned sequence fragments from the A genome (G. arboreum var. YZ) and the Dt genome (G. hirsutum var. TM-1) corresponding to the D genome (G. raimondii) indicated by two black arrows in A. Amplification of the At genomic fragment from G. hirsutum failed probably due to sequence difference between the At and Dt genome at the primer annealing regions. miRCandidate1 (miRCan1) and miR482e.2 are highlighted in blue and red, respectively. Three SNPs that abolished the production of miRCan1 in the A genome are boxed.