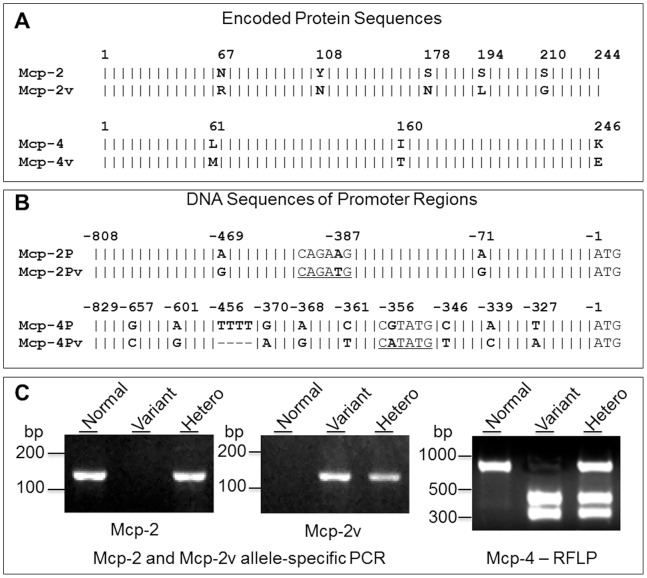
Figure 2. Identification of Mcp-2 and Mcp-4 gene variants.
Schematic alignment of amino acid sequences (A.) and promoter region DNA sequences (B.) of Mcp-2P and Mcp-4P from control B6 and variant B6-cma mice. Variant amino acid residues and nucleotide bases are highlighted in bold. A vertical line “|” denotes identical amino acids or bases, and a dash “–” stands for deletions. Putative Mitf binding consensus motifs (CANNTG E-boxes) and an NdeI restriction cleavage site (CATATG) in the variant form of Mcp-2 or Mcp-4 are underlined. C. Detection of Mcp-2 and Mcp-4 gene variations in the promoter regions by allele-specific PCR and NdeI restriction fragment length polymorphism (RFLP), respectively. Normal form of Mcp-2P was detected by PCR with primers 5′-ctcacactggtcaacacaaacatta and 5′-tctgctgttaaacacaaacacagtct, while the Mcp-2P variant was amplified by using primers 5′-ctcacactggtcaacacaaacattg and 5′-tctgctgttaaacacaaacacagtca. The expected product size for both is 131 bp. The variant form of Mcp-4P was revealed by NdeI digestion which gave rise to two fragments while the normal form of Mcp-4P was not cleaved. Data show results for both homozygous and heterozygous mice.