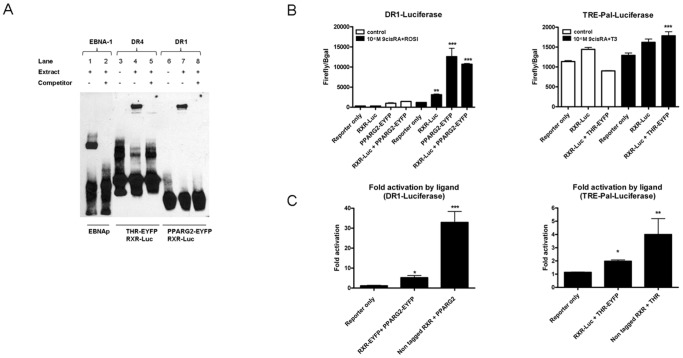
Figure 2. Functional validation of NR fusion proteins.
(A) Electrophoretic mobility shift assay (EMSA) experiments with NR fusion proteins encoding RXR-Luc, THR-EYFP and PPARG2-EYFP. Nuclear extracts were prepared from HEK293T cells co-transfected with RXR-Luc and THR-EYFP or RXR-Luc and PPARG2-EYFP encoding plasmids. Several biotinylated RE oligonucleotides were mixed with nuclear extracts containing overexpressed NR fusion proteins: A DR4 probe was used to test the DNA binding of RXR-Luc+THR-EYFP (lane 4), and a DR1 probe was used for RXR-Luc+PPARG2-EYFP (lane 7). In the absence of nuclear extracts no signal was obtained (lanes 3 and 6). As a positive control for EMSA, a biotin-labeled 60 bp duplex bearing the Epstein-Barr Nuclear antigen 1 (EBNA-1) binding sequence was incubated with an extract containing the EBNA protein (lane 1). For each condition, the specificity of the gel shift experiment was determined by the addition of a 200-fold excess of the corresponding unlabeled double stranded consensus sequence (lanes 2, 5 and 8). (B) Histograms represent transcriptional activity of NR fusion proteins RXR-Luc, PPARG2-EYFP (left) or RXR-Luc and THR-EYFP (right) monitored with gene reporters containing DR1 responsive elements or thyroid responsive element palindomic (TREPAL) in their promoters: Hela cells were transiently transfected with RXR-Luc and PPARG2-EYFP or RXR-Luc and THR-EYFP expression plasmids together with a PPAR DR1 (left) or a TREPAL firefly luciferase gene reporter construct (right), respectively. A pCMV-β-galactosidase plasmid was used for normalization. 6 hours after transfection some cells were stimulated 24 h with 10−6 M rosiglitazone (ROSI) or 10−6 M of triodothyronine (T3) and 10−6 M of 9cis retinoic acid (9cis RA). After cells lysis, transcriptional activity was measured by monitoring firefly luciferase activities normalized by β-galactosidase activities. (C) Histograms represent transcriptional fold activation by ligand of transfected tagged constructs, RXR-Luc and PPARG2-EYFP (left) or RXR-Luc and THR-EYFP (right) compared to non tagged plasmids. Values shown are mean ± SD. (n = 2–3). Statistical differences were analyzed by one-way ANOVA followed by Bonferroni’s post hoc test: *P<0.05; **P<0.01; ***P<0.001.