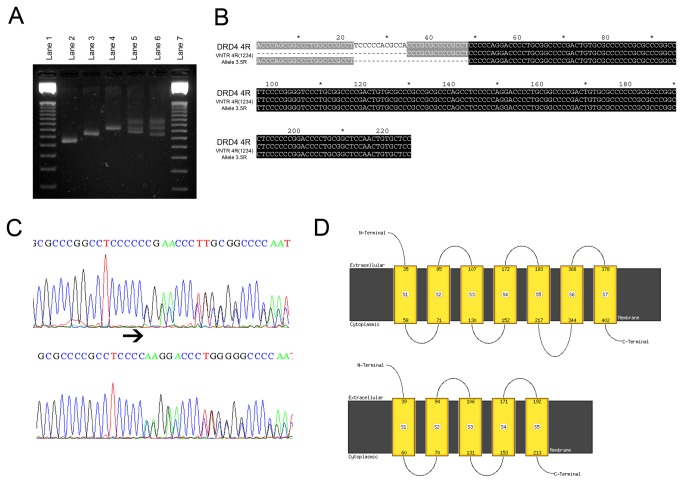
Figure 2. Characterization of DRD4 novel deletions.
(A) PCR analysis of 3.5 allele. Lane 1: DNA 50bp marker, Lane 2: homozygote for 2 repeats; Lane 3 homozygote for 3 repeats; Lane 4: homozygote for 4 repeats; Lane 5: heterozygote for 3 and 4 repeats; Lane 6: heterozygote for 3.5 and 4 repeats. PCR products were run on a 3.5% agarose gel. A heteroduplex band is seen for heterozygotes in lanes 5 and 6. (B) The allele 3.5R alignment. The 3.5R refers to an allele with a length between 3 and 4 repeats. Sequencing of this size allele revealed a deletion of 26bp sequence (11bp immediate 5’ downstream sequence of the VNTR and 15bp inside the first repeat of VNTR) of the most common 4R haplotype. (C) Sequence chromatogram of DRD4-7R nucleotide deletions(arrow) at VNTR positions117 (del117C, above) and 20 (del20C, below). (D) Topology prediction of proteins encoded by VNTR with (below) and without (above) deletions. Schematic diagram of the transmembrane region (yellow). The deletions exclude the prediction of the two last transmembrane domains.