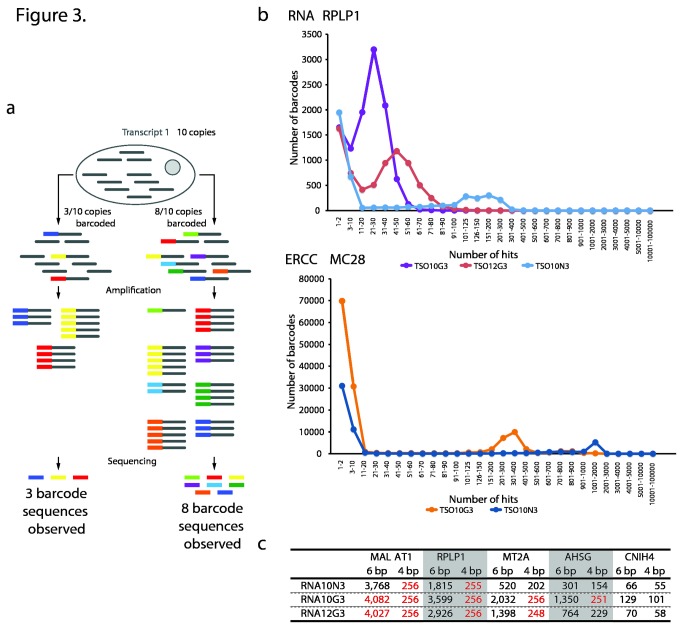
Figure 3. | UMI length.
(a) shows the UMI principle and the output depending on reaction efficiency. If three out of the ten transcript molecules in total are labeled with the unique identifier (i.e. barcoded), only three barcodes will be observed in the sequencing data, translating into a transcript count of three for this particular transcript. Converting eight out of the ten molecules leads to the identification of eight barcodes in the sequencing reads. A UMI can become saturated if the number of transcripts copies exceeds the number of possible UMI combinations.(b) shows the distribution of barcodes for RPLP1 and spike MC28 in the performed reactions. Corresponding graphs for the other investigated transcripts are shown in Figure S6 and Figure S7 in File S1 for the analyzed RNA spikes. The reactions with the N10rG3 TSOs exhibited the highest complexity, followed by the reactions employing the N12rG3 and, lastly, the N10rN3 oligonucleotides. In (c) 6-base and 4-base barcodes were extracted from the MALAT1, RPLP1, MT2A, AHSG and CNIH4 reads. The red numbers indicate that the UMI has become saturated.