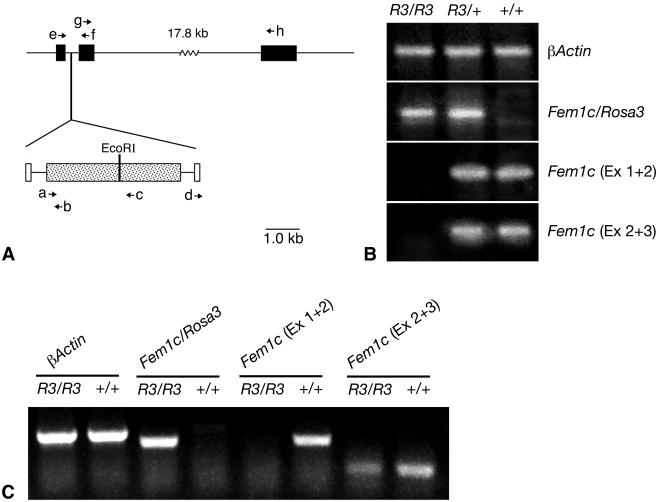
FIG. 6.
βGeo insertion disrupts normal Fem1c expression. (A) Map of Fem1c on chromosome 18 showing the insertion site of the βGeo trap vector. Exons of Fem1c are shown as filled boxes. The βGeo coding region is shown as a stippled box, and the flanking retroviral long terminal repeats are shown as open boxes. βGeo contains an internal EcoRI restriction site that was used for inverse PCR experiments. The relative positions of various primers (a to h) used for PCR experiments are shown with arrows. The sequences and names of these primers are detailed in Table 1. βGeo is inserted into the first intron of the Fem1c gene. (B) Ethidium bromide-stained agarose gels showing the results of RT-PCR analysis of transcripts from the Fem1c alleles. Total RNA was isolated from Rosa3 mouse brains and used to make first-strand cDNA. The genotypes of the mice are indicated above each lane. β-Actin amplification was used as a control for equivalent amounts of template cDNA (top panel). The results of RT-PCR with primers e and b (BF4.1 and LacZ.2, respectively) show that the βGeo coding region is spliced onto exon 1 of Fem1c only in R3/R3 and R3/+ mice (second panel from top). Sequence analysis of this fragment shows correct splicing at the end of exon 1. The results of RT-PCR with primers e and f (BF4.1 and BF4.2, respectively) indicate appropriate splicing of exons 1 and 2 (Ex 1 + 2) of Fem1c in mice that carry one or both wild-type alleles. No splice was evident in R3/R3 mice (third panel). The results of RT-PCR with primers g and h (BF4.6 and BF4.7, respectively) indicate appropriate splicing of exons 2 and 3 of Fem1c. No splice that bypasses exon 1 is detected in R3/R3 mouse brain under these PCR conditions (30 cycles) (fourth panel). These data show that Fem1c expression is knocked out in R3/R3 mice. Ex 2 + 3, exons 2 and 3. (C) Ethidium bromide-stained agarose gel of RT-PCR products generated from extended (40-cycle) PCR. All products were generated from testes RNA isolated from R3/R3 or wild-type mice. Samples from R3/R3 mice exhibit a faint band indicative of a transcript containing correctly spliced exons 2 and 3 of Fem1c. No transcript containing exons 1 and 2 of Fem1c in the same mice was detected. Primers used for these products were the same as those used as describe for panel B, and β-actin was used as a control.