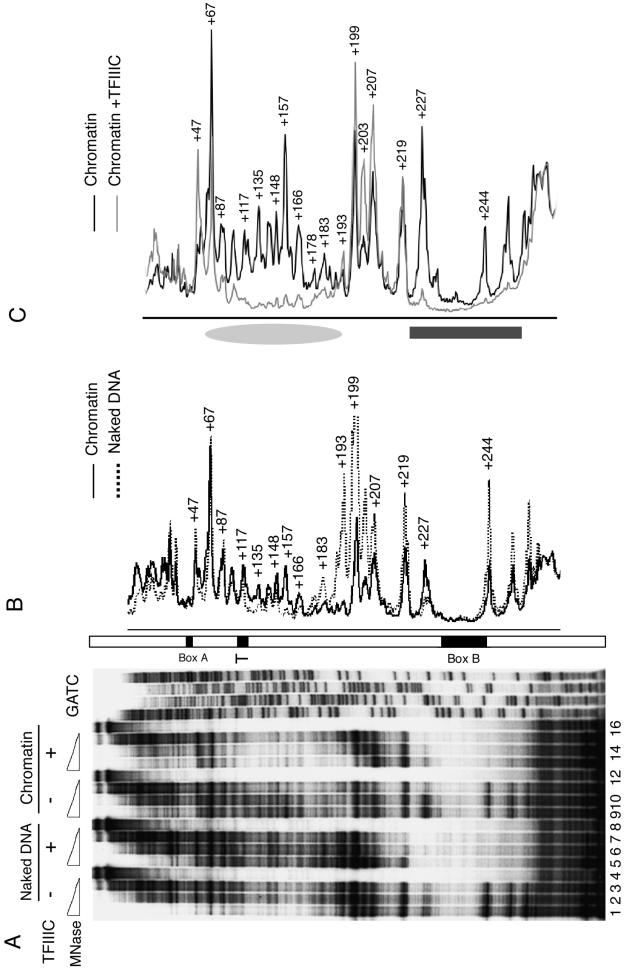
FIG. 3.
Analysis of chromatin remodeling by MNase digestion. (A) Higher-resolution footprinting. Chromatin without or with an 11-fold molar excess of TFIIIC added at the start of assembly (lanes 9 to 16) was subjected to three levels of MNase digestion and compared with naked DNA without (lanes 1 to 3) or with (lanes 5 to 7) TFIIIC. Lanes 4, 8, 12, and 16 show primer extension on undigested DNA. The 5′ end of the primer extension probe was 31 bp away from the 3′ end of box B, complementary to the U6 gene top strand. The GATC ladder was generated with the same primer. Positions of box B, the transcription terminator (T), and box A are marked on an open bar. (B and C) A nucleosome-size footprint on chromatin in the presence of TFIIIC. Aligned digestion profiles are taken from panel A. (B) Naked DNA (lane 2) and chromatin (lane 10) without TFIIIC. (C) Chromatin without TFIIIC (lane 9) and with TFIIIC (lane 14). Numbers denote positions of MNase cuts with reference to the transcriptional initiation site; +244 denotes the single cut in box B. Protection in the region of box B is marked with a dark gray box, while the gray ellipse marks the position of nucleosomal protection in the presence of TFIIIC.