Figure 4.
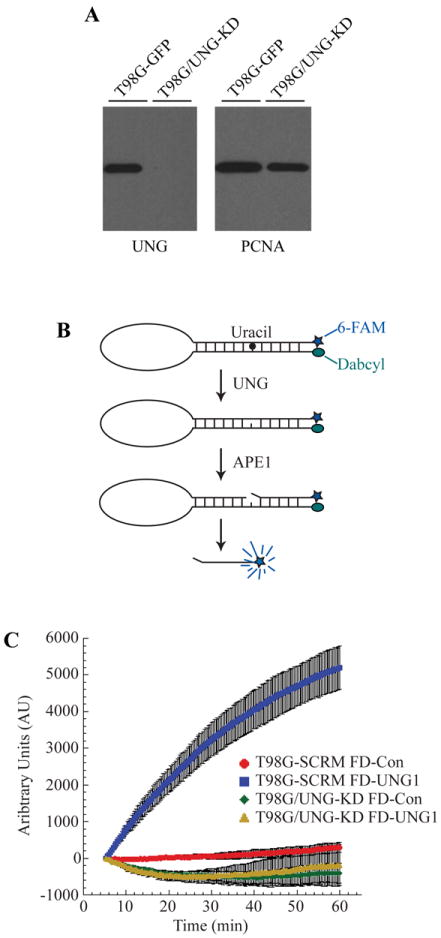
Depletion of UNG protein and uracil glycosylase activity in glioma cells. (A) Knockdown of UNG mRNA corresponds with decreased UNG protein levels. Control and UNG-KD cell line nuclear extracts were resolved in a 4-20% SDS/PAGE gel and immunoblotted for UNG. The blot was stripped and re-probed for PCNA, which was used as a loading control. (B) Molecular beacon model for real-time detection of uracil removal at 37°C in nuclear extracts. (C) Knockdown of UNG abolishes ability to remove uracil from DNA as determined by DNA Glycosylase Molecular Beacon Activity Assay. Specific activity of DNA glycosylase activity for the uracil lesion was measured in nuclear extracts from T98G-SCR (control-beacon, red circles; uracil-beacon, blue squares) and T98G/UNG-KD (control-beacon, green diamonds; uracil-beacon, yellow triangles). The mean fluorescence response unit of three experiments is plotted ± SEM.
