Abstract
Because activation of p53 can trigger cell cycle arrest and apoptosis, it is necessary for a cell to suppress this activation until it is absolutely required for survival. The mechanisms underlying this important regulatory event are poorly understood. Here we show that nucleophosmin (NPM) acts as a natural repressor of p53 by setting a threshold for p53 activation in response to UV radiation. NPM binds to the p53 N terminus and inhibits p53 transcriptional activity by more than 70%. Our data indicate that the levels of NPM in a cell determine the UV dose at which the tumor suppressor p53 can be phosphorylated on Ser15. Moreover, we show that NPM is a substrate for the UV-activated protein kinase ATR and inhibits the UV-induced p53 phosphorylation at Ser15. In addition, NPM forms a complex with p53 and ATR in vivo. These data suggest that NPM is an early responder to DNA damage that prevents premature activation of p53. In normal cells, NPM could contribute to suppressing p53 activation until its functions are absolutely required while in cancer cells overexpression of NPM could contribute to p53 inactivation and tumor progression.
Activation of the tumor suppressor p53 in response to DNA damage is an important event that prevents a normal cell from undergoing cellular aberrations that can lead to cancer progression. Because p53 can trigger cell cycle arrest or apoptosis, it is fundamental that the activation of p53 remains in check until it is absolutely required for cellular homeostasis. The restraint on p53 must be sufficient to allow normal growth and development while allowing it to retain the capacity for rapid induction in response to stress associated with tumor progression (28). In recent years, much emphasis has been put on the mechanisms that activate p53 in response to genotoxic stress, but little is known about how p53 functions are kept on hold under normal or low-stress conditions. Here we show data indicating that nucleophosmin (NPM) is a natural repressor of p53 that sets a threshold for p53 response to UV radiation.
NPM, also known as B23, NO38, and numatrin (30), is a nucleolar protein that was initially identified as an important player in ribosome biogenesis (5). Since then a number of cellular activities associated with NPM indicate that the protein has multiple functions, especially in cell proliferation. For example, in anaplastic large-cell lymphoma NPM is fused to a receptor tyrosine kinase (anaplastic lymphoma kinase [ALK]) and works as an oncogene (10). NPM protein levels are 20 times higher in Novikoff hepatoma and 5 times higher in hypertrophic rat liver than in normal rat liver (5). NPM binds to pRb and synergistically stimulates DNA polymerase α (25). NPM also binds to interferon regulatory factor 1 (IRF-1) and inhibits its tumor suppression function, probably by preventing expression of p21 (19). Another indication of NPM's role in cell proliferation is its association with the nucleolar organizer regions. The nucleolar organizer regions correlate with cell proliferation and tumor progression (9). All these data suggest that NPM plays an important role in cell proliferation.
Nonetheless, a recent report (7) indicates that NPM stabilizes p53 in response to DNA damage. However, it is not clear how these data can be reconciled with the fact that NPM is overexpressed in cancer cells (5), is associated with cell proliferation and tumor progression (9), and has an effect opposite that of p53 on centrosome duplication (22).
Our data indicate that NPM interacts with p53 and down regulates its transcriptional activity by more than 70%. NPM binds to the p53 N-terminal end and prevents p53 phosphorylation at Ser15 in response to low doses of UV radiation. In addition, down regulation of NPM by small interfering RNA (siRNA) allowed p53 phosphorylation to occur at a lower dose of UV radiation. These data suggest that NPM sets a threshold for p53 activation by DNA damage. NPM could thus repress p53 activation until the levels of DNA damage require it. Activation of p53 at an inappropriate time could compromise cellular growth and even trigger apoptosis.
MATERIALS AND METHODS
Cell culture and treatments.
Human large-cell lung carcinoma H1299 cells were purchased from the American Type Culture Collection (Manassas, Va.). Human colorectal carcinoma RKO cells were provided by A. J. Fornace, Jr. (National Cancer Institute, National Institutes of Health, Bethesda, Md.). The cells were grown in RPMI 1640 containing 10% fetal bovine serum (Invitrogen Corp., Carlsbad, Calif.). Treatments of the cells were as described previously (12) except that the UV source was a Philips 30-W germicidal lamp. Intensities of the UV lamp were determined with a UVX radiometer (UVP Inc., Upland, Calif.). The cells were harvested 4 h after treatment and processed as described elsewhere in the text. Fluorescence-activated cell sorter analysis was performed on RKO cells transfected with siRNA (12 nM) and exposed to UV radiation (10 J m−2) 24 h after transfection. The cells were harvested and fixed 12 h after irradiation, and the DNA was stained with propidium iodide (0.4 mg/ml; Roche, Indianapolis, Ind.). The cell cycle distribution was analyzed on a BD Bioscience cell sorter (Palo Alto, Calif.).
Plasmids and chloramphenicol acetyltransferase (CAT) assay.
The H1299 cells were transfected with either pCMV3 empty vector (Stratagene, La Jolla, Calif.), pCMV-NPM, pCMV-p53, pG13 CAT, or the p53REX vector. The pCMV-p53 and pG13 CAT vectors were provided by Bert Vogelstein (Johns Hopkins University, Baltimore, Md.). The full-length (FL) human NPM cDNA (Pui-Kwong Chan, Baylor College of Medicine, Houston, Tex.) was amplified by PCR and cloned into the BamHI/XhoI sites of the pCMV.3 vector. The p53REX vector was provided by Al Fornace, Jr. This vector contains five p53 binding sites identical to the p53 binding site in the GADD45 third intron except that the last purine, G, is replaced by C (26). The RKO cells were transiently transfected with pG13 CAT and NPM expression vectors and exposed to UV radiation (20 J m−2) 24 h after transfection. All cells were transfected using FuGENE reagent (Roche Molecular Biochemicals) according to the manufacturer's recommendations. Cells were harvested and lysed 48 h after transfection, and the protein concentration and CAT activity were measured as described before (20).
GST pull-down assays.
The glutathione S-transferase (GST) FL p53 expression vector was constructed by PCR amplification with a forward primer, 5′-CCGCGTGGATCCATGGAGGAGCCGCAG-3′, and a reverse primer, 5′-GCCGCTCGAGTCTCAGTCTGAGTCAGGCCC-3′. The amplified DNA was cloned into the pGEX-6P-2 vector (Amersham Pharmacia Biotech, Piscataway, N.J.). The p53 DNA binding domain (DBD) was amplified using the same forward primer as for the FL vector but with the sequence 5′-GCCGCTCGAGTCTCATTTCTTGCGGAGATT3′ as a reverse primer and was cloned into pGEX-dT (Amersham). The p53 N-terminal region (NT) was amplified using the same forward primer as for the FL vector but with the sequence 5′-GCCGCTCGAGTCTCAGGTTTTCTGGGAAGG-3′ as a reverse primer and was cloned into the same vector as was DBD. All constructs and plasmids were cut with BamHI (5′ end) and XhoI (3′ end) (New England Biolabs, Beverly, Mass.). Proteins were expressed and purified as recommended by the manufacturer (Amersham). The pull-down assays were performed as described in reference 23 with the exception that the GST-p53 proteins were purified first before being incubated with the NPM bacterial extracts. The GST-p53 proteins (300 ng) and 10 μl (100 μg) of BL-21 bacterial extracts expressing recombinant NPM (31) were incubated for 1 h at 4°C, and then the glutathione Sepharose beads were added and the incubation was continued overnight at 4°C with gentle mixing. Mixtures were spun down, and the pellets were washed six times with RIPA buffer (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 0.5% sodium desoxycholate, 1% NP-40, and 0.1% sodium dodecyl sulfate [SDS]). The samples were resolved on an SDS-12% polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was analyzed by Western blotting with NPM polyclonal antibody (Ab) (31).
siRNA.
The 21-nucleotide (nt) double-stranded RNA was synthesized by Xeragon Inc. (Huntsville, Ala.). The sequence (5′-CCACAGAAAAAAGUAAAAC-3′) corresponds to 19 nt from the NPM open reading frame (nt 547 to 565) and a two-dT overhang. The RNA (12 nM) was added to the cells along with 2 μg of pCMV.3 empty vector (Stratagene). The scrambled RNA is composed of four random sequences of 19 nt with a two-UU overhang on each side (nonspecific control duplexes XIII, pool of four; Dharmacon, Inc., Boulder, Colo.). The siRNAs were transfected in RKO cells with FuGENE reagent according to the manufacturer's recommendations. The cells were exposed to UV radiation 24 h after transfection.
Abs and Western blots.
The polyclonal Ab to NPM was generated in rabbits immunized with a synthetic peptide (residues 221 to 238) as the antigen (31). Cellular extracts were harvested, separated on an SDS-12% polyacrylamide gel, and transferred to a nitrocellulose membrane. The p53 DO-1 monoclonal Ab was diluted 1/1,000 (Calbiochem, San Diego, Calif.), the p53 Ser15 phosphospecific polyclonal Ab (Cell Signaling, Beverly, Mass.) was diluted 1/500, and the p21 polyclonal Ab (BD PharMingen, Heidelberg, Germany) was diluted 1/500. Western blotting was performed with the respective secondary Ab conjugated to horseradish peroxidase and reacted with a chemiluminescent substrate (ECL; Amersham) according to the manufacturer's recommendations. Fold induction was calculated by densitometry and normalized to actin.
Coimmunoprecipitation.
Extracts of RKO cells exposed or not to 14 J of UV radiation m−2 were prepared by growing 106 cells and harvesting them 4 h after treatments. The cells were lysed in RIPA buffer. The protein extracts (7.5 to 12 mg) were incubated with 15 μl of either Ser15 p53 Ab or DO-1 Ab at room temperature. Protein A agarose beads (20 μl; Santa Cruz Biotechnology, Santa Cruz, Calif.) were then added, and the samples were incubated overnight at 4°C. The beads were spun down, washed six times with RIPA buffer, and loaded on an SDS-12% polyacrylamide gel. The samples were transferred to a nitrocellulose membrane and incubated with NPM Ab as described above.
Kinase assay.
The Flag-ATR construct containing the FL cDNA clone from human ATR in pcDNA3 (Invitrogen) was provided by A. Sancar (University of North Carolina, Chapel Hill). The Flag-ATM containing the FL cDNA clone from the human ATM in pcDNA3 (3) was provided by Mike B. Kastan (St. Jude Children's Research Hospital, Memphis, Tenn.). H1299 cells were transiently transfected as described above with the Flag-ATR or Flag-ATM plasmid. Forty-eight hours after transfection, the cells were exposed to either 10 or 14 J of radiation m−2 (Flag-ATR) or 15 μg of bleomycin (Sigma, St. Louis, Mo.)/ml (Flag-ATM), and the cells were harvested 4 h later. The Flag proteins were purified with a Flag immunoaffinity kit according to the protocol of the manufacturer (Sigma). The kinase reaction was performed with 500 ng of Flag-ATR or Flag-ATM, 300 ng of recombinant NPM, and 2 mCi of [γ-32P]ATP in 30 μl of 10 mM Tris-HCl (pH 7.4)-150 mM NaCl-10 mM MgCl2-0.5 mM dithiothreitol (kinase reaction buffer) for 30 min at 37°C. Samples were run on an SDS-12% polyacrylamide gel. The gel was dried and exposed to X-ray-sensitive film. Where indicated, wortmannin (Sigma) was incubated at 30°C with the kinase for 30 min prior to the addition of the substrates. In the case of caffeine (Sigma), the indicated concentrations were added just before the addition of the substrates. In the DNase I sample, the enzyme was incubated with the kinase at 30°C for 30 min before the addition of the substrate and caffeine.
RESULTS AND DISCUSSION
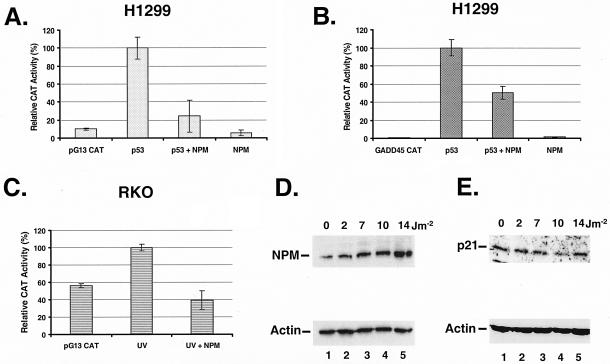
The identification of NPM as a stress-activated protein (31) and its inhibitory effect on IRF-1 (19), a transcription factor regulating the cyclin-dependent kinase inhibitor p21, led us to investigate NPM's potential effect on p53, another regulator of p21 (11). We first performed transient-cotransfection assays with a p53 reporter gene, pG13 CAT (18), and expression vectors for p53 and NPM in the human large-cell lung carcinoma line H1299. These cells have a p53-null genotype (17) that facilitates the interpretation of the results due to the absence of endogenous p53 protein. Our data (Fig. 1A) indicate that NPM inhibits p53 transcriptional activity by more than 70%. The levels of inhibition are comparable to those that NPM imposed on the promoter of IRF-1, another tumor suppressor gene (19). NPM inhibition is specific and not due to a general repressor effect, since NPM is known to have no effect on a β-actin promoter (19) and activates a PCNA promoter (30). The inhibitory effect of NPM on at least two tumor suppressors may thus explain its oncogenic transformation activity in NIH 3T3 cells (19).
FIG. 1.
(A) NPM represses p53 transcriptional activity. For the CAT assay, H1299 cells were transfected with the p53 reporter gene construct pG13 CAT and either NPM, p53, or both. The empty pCMV plasmid was used to normalize the plasmid content (6 μg) in all transfections. The cells were harvested 48 h after transfection. The relative CAT activity is expressed as a percentage of the total activity obtained with p53 and pG13 CAT (p53). The data represent the averages of two experiments performed in duplicate, and the error bars correspond to standard errors of the means. (B) Same as panel A except that the cells were transiently transfected with a reporter gene containing five repeats of the p53 binding site from the GADD45 third intron (GADD45 CAT). (C) RKO cells were transiently transfected with pG13 CAT and where indicated with NPM. The cells were exposed to UV radiation (20 J m−2) as indicated. (D and E) Up regulation of NPM correlates with p21 down regulation by UV radiation. For Western blot analyses, RKO cells were exposed to the indicated doses of UV radiation and harvested 4 h later. Total cellular protein (40 μg for NPM or 200 μg for p21) was reacted with NPM Ab (D) or p21 Ab (E). Blots were reacted with actin Ab for loading control.
In order to evaluate NPM's effect on a bona fide p53 binding site, we repeated the CAT assay with a reporter gene construct containing five repeats of the p53 binding site of the GADD45 third intron (17). The data shown in Fig. 1B indicate that similar results were obtained with this reporter where NPM inhibited more than 50% of the CAT activity. We then proceed to determine whether NPM could inhibit the transcriptional activity of endogenous p53. We transiently transfected the pG13 CAT vector in colon carcinoma RKO cells and used UV radiation to induce endogenous p53 levels. The RKO cells have a wild-type p53 genotype (17) and relatively high basal p53 levels (32). The data shown in Fig. 1C indicate that basal p53 levels are sufficient to activate the p53 reporter gene but that UV radiation can increase the basal activity by almost twofold. Overexpression of NPM reduced the UV-induced p53 transcriptional activity by more than 50%. These data suggest that NPM could repress p53 endogenous transcriptional activity on its downstream effector genes.
To evaluate the potential effect of NPM on the p53 response in vivo, we measured the response of cellular NPM to p21, a p53 downstream effector gene (11), after UV radiation. The data presented in Fig. 1D indicate that, as previously reported (31), NPM protein levels increased in a dose-dependent manner in response to UV radiation. Interestingly, p21 protein levels decreased as NPM protein levels increased from 2 to 10 J of UV radiation m−2 (Fig. 1E, lanes 1 to 4). At 14 J m−2, NPM protein levels continued to increase (Fig. 1D, lane 5) while p21 protein levels stopped decreasing and returned to basal levels or higher (Fig. 1E, lane 5). The down regulation of p21 protein levels in response to low doses of UV radiation is not without precedent; at least two other groups have reported similar results in several cell lines including RKO cells (29). The inverse correlation between NPM and p21 levels at low but not high doses of UV radiation suggests that NPM may be involved in an early sensor mechanism to regulate p21 expression in response to genotoxic stress.
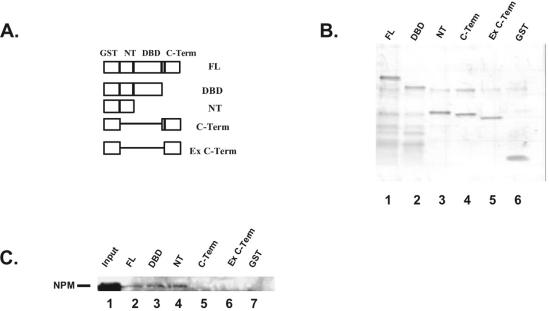
NPM binds directly to IRF-1 to mediate its repressive activity (19). A recent report has shown that NPM also binds directly to p53 (7). The NPM domain that interacts with p53 (amino acids 187 to 295) (7) overlaps with the domain that is phosphorylated by CDK2/cyclin E and controls centromere duplication (22). To determine which p53 domain is involved in the NPM interaction, we engineered several overlapping p53 deletion mutants fused to GST (Fig. 2A). Several functional domains have been described for p53. The p53 N-terminal transactivation domain (NT) interacts with components of the transcriptional machinery such as CBP/p300 and with MDM2, which targets p53 for degradation (28). The NT is also the site of phosphorylation for several stress-activated kinases such as ATM and ATR. The central-sequence-specific DBD is where most of the point mutations found in cancer cells occur. The C-terminal domain (C-term) is composed of the tetramerization domain, responsible for p53 oligomerization, and the extreme C-terminal domain that acts as a negative regulator. Incubation of recombinant NPM with the p53 fusion proteins indicates that NPM binds to all constructs containing the NT but not to the constructs lacking it. These data indicate that the p53 C-terminal domains are not required for the interaction and that binding to p53 NT is sufficient to form a complex (Fig. 2C). Coomassie blue staining of an SDS-polyacrylamide gel indicates that approximately the same amount of protein was used in each reaction (Fig. 2B).
FIG. 2.
NPM interacts with p53 N-terminal end. (A) The different domains of p53 were amplified by PCR. (B) SDS-polyacrylamide gel electrophoresis of GST-p53 deletion mutants. (C) Western blot analysis. The p53 fusion proteins were incubated with NPM bacterial extracts. The eluted proteins were reacted with NPM Ab.
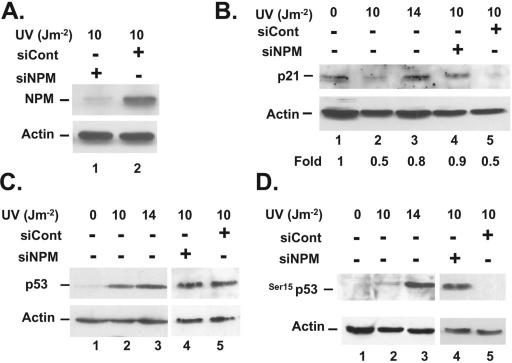
Our data (Fig. 1D and E) indicate that NPM levels inversely correlate with p21 protein levels in response to low doses of UV radiation. In addition, our data (Fig. 1A to C) indicate that NPM can down regulate p53 transcriptional activity. To verify whether NPM is directly involved in p21 down regulation in response to UV radiation, we down regulated NPM protein levels by the siRNA method. As shown in Fig. 3A, NPM siRNA (siNPM) reduced the levels of NPM by at least 80% while a control scrambled RNA did not affect NPM levels. In the absence of siNPM, the expected decrease in p21 protein levels was observed at 10 J m−2 (Fig. 3B, lane 2). At 14 J m−2, p21 levels increased again, but when the cells were exposed to 10 J of UV m−2 in the presence of siNPM, p21 protein levels remained at basal levels (Fig. 3B, lane 4). Exposing the cells to 10 J m−2 in the presence of a control scrambled siRNA did not increase p21 levels. These data indicate that NPM directly affects the down regulation of p21 observed in response to low doses of UV radiation.
FIG. 3.
NPM sets a threshold for p53 response to UV radiation. The figure shows results of Western blot analyses. (A) RKO cells treated with siNPM or control scrambled RNA (siCont) were exposed to UV radiation and harvested. The proteins (30 μg) were reacted with NPM Ab. (B) Same as panel A, but the cells were exposed to the indicated dose of UV radiation and 200 μg of protein was loaded before the blot was reacted with p21 Ab. (C) Same as panel B, but the proteins (200 μg) were reacted with p53 Ab. (D) Same as panel B, except that the blot was reacted with a p53 Ab specific for phosphorylation at Ser15. All blots were reacted with actin Ab for a loading control.
We then asked whether the increase in p21 levels observed at 10 J m−2 in the presence of siNPM (Fig. 3B) was p53 dependent. The data shown in Fig. 3C indicate that, in the absence of siRNA (lanes 1 to 3), p53 levels increased in response to UV radiation in a dose-dependent manner. Addition of siNPM or a control scrambled siRNA (lanes 4 and 5) did not affect p53 overall protein levels when the cells were exposed to 10 J of UV radiation m−2 (compare lane 2 with lanes 4 and 5). Because NPM is induced by UV radiation (Fig. 1D) and interacts with the p53 N-terminal end (Fig. 2B) and p53 is phosphorylated at Ser15 in response to UV radiation (2), we then evaluated the effect of NPM on p53 phosphorylation at Ser15 in response to UV radiation. Phosphorylation of p53 at Ser15 is important for p53 stability and function in response to genotoxic stress (2). Our data indicate that a dose of 14 J m−2 is required to generate a substantial amount of p53 phosphorylated at Ser15 in vivo (Fig. 3D, lane 3). However, when the levels of NPM are reduced by siRNA, the threshold to induce significant phosphorylation of p53 at Ser15 is lowered to 10 J m−2 (Fig. 3D, lane 4). This effect is specific because it was not observed when a scrambled siRNA was transfected in the cells (Fig. 3D, lane 5).
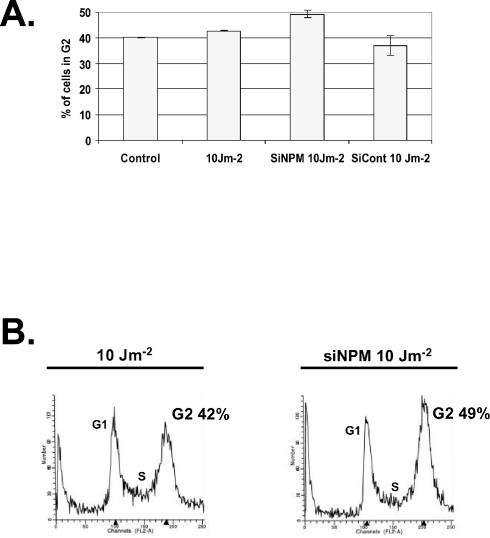
To determine whether activation of p53 at a lower UV dose and the subsequent induction of p21 had any functional significance, we evaluated the effect of NPM down regulation on cell cycle distribution following exposure to 10 J of UV radiation m−2. The data shown in Fig. 4 indicate that a subtle but significant increase was observed in the number of siNPM-RKO cells arrested in G2 following exposure to UV radiation. The increase in G2 (7%, Fig. 4B) correlates with the increased p53 phosphorylation and the increased p21 expression observed with reduced levels of NPM (Fig. 3). This relatively small effect is significant because it was not observed with the control scrambled RNA (Fig. 4A). The overall NPM effect on cell cycle progression is probably even more pronounced than what was observed here, since only a fraction of the cells incorporate the siNPM during the transient transfection. Our data are in good agreement with a previous report (14) showing that down regulation of NPM by antisense transfection results in a delay of cell entry into mitosis and consequently increases the number of cells in G2. The role of p21 in the UV-induced G2 arrest is believed to be p53 dependent and to affect the late (prolonged) G2 arrest (21).
FIG. 4.
Down regulation of NPM alters cell cycle distribution of UV-treated cells. (A) Fluorescence-activated cell sorter analysis. RKO cells were left untreated (control) or transfected with siNPM or scrambled RNA (siCont) and where indicated exposed to UV radiation (10 J m−2). Percentages of cells in G2 are as indicated and represent the averages of at least three independent experiments. (B) Cell cycle distribution profiles. Percentages of cells in G2 are as indicated.
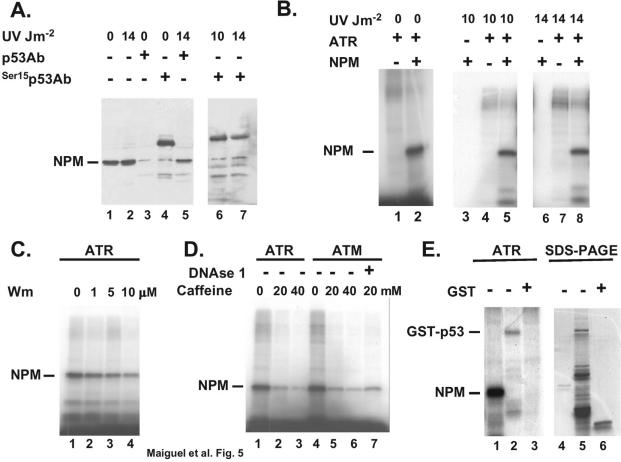
There are at least two mechanisms by which NPM could set a threshold for p53 phosphorylation at Ser15. The first would be if NPM associates with p53 when p53 is underphosphorylated (<10 J m−2) but dissociates from this site when Ser15 is hyperphosphorylated (>14 J m−2). To test this possibility, we performed immunoprecipitation with protein extracted from RKO cells exposed or not to 14 J of UV radiation m−2. The data shown in Fig. 5A indicate that, in the absence of UV radiation (lanes 3 and 4), the p53 and the Ser15 phosphospecific p53 Abs can immunoprecipitate a small amount of NPM from cellular extracts (7.5 to 12 mg). The NPM interaction with p53 has also been observed previously with immunoprecipitation performed with NPM Ab (7). However, when cells are exposed to UV radiation (10 and 14 J m−2), the p53 Ab as well as the phosphospecific p53 Ab can still immunoprecipitate NPM (Fig. 5A, lanes 5 to 7). Therefore, phosphorylation of p53 at Ser15 does not abolish interaction with NPM. However, the amounts of NPM that associate with p53 phosphorylated at Ser15 were approximately the same under all conditions, while the amounts of NPM that associate with p53 increased at 14 J m−2 (lane 5). This may reflect the fact that, in general, cells contain more p53 than p53 phosphorylated at Ser15 (15, 24) or that phosphorylation at Ser15 contributes partially to reduce the interaction of p53 with NPM.
FIG. 5.
NPM interacts with p53 phosphorylated at Ser15 and is an ATR substrate. (A) Immunoprecipitation of NPM (lanes 3 to 7) from RKO protein extracts (7.5 to 12 mg) with p53 Ab (DO-1) or the phosphospecific Ser15 p53 Ab (Ser15p53Ab). The doses of UV radiation are indicated. The levels of NPM present in the extracts before immunoprecipitation (∼1% of the input) are shown (lanes 1 and 2). (B) Phosphorylation of recombinant NPM with Flag-ATR immunoprecipitated from H1299 cells exposed to the indicated dose of UV radiation. (C) Inhibition of ATR by wortmannin (Wm). Phosphorylation was performed as in panel B, with the exception that the immunoprecipitated kinase was incubated with the indicted concentration of wortmannin for 30 min prior to the addition of NPM. (D) Phosphorylation (lanes 1 to 3) was performed as in panel B, with the exception that the indicated concentration of caffeine was added to the reaction mixture prior to the addition of NPM. Phosphorylation with the ATM kinase (lanes 4 to 7) was performed as described in Materials and Methods, and the indicated concentration of caffeine was added prior to the addition of NPM. DNase I (lane 7) was incubated with the immunoprecipitated kinase for 30 min prior to the addition of NPM. (E) Phosphorylation of recombinant NPM and GST-p53 by ATR was performed as in panel B. Purified GST is used as a negative control (lane 3). Results for SDS-polyacrylamide gel electrophoresis (PAGE) of the recombinant proteins used in the kinase assay are shown (lanes 4 to 6).
Because phosphorylation of p53 at Ser15 did not completely abolish NPM binding (Fig. 5A, lanes 6 and 7), we also considered the possibility that NPM could compete with p53 for the kinase responsible for phosphorylation at this site. Phosphorylation of p53 at Ser15 in response to UV radiation is mediated by the ATR kinase (28). We first aimed at determining whether NPM is a substrate for ATR. The data shown in Fig. 5B indicate that NPM is a good substrate for ATR immunoprecipitated from cells exposed to either 0, 10, or 14 J of UV radiation m−2 (lanes 5 and 8). In contrast to ATM, ATR activity does not increase following UV radiation, but rather the kinase relocates to DNA damage-induced nuclear foci (1). In accordance with this phenomenon we found that NPM was phosphorylated by Flag-ATR even in the absence of UV radiation (Fig. 5B). Because NPM is a shuttle protein, proximity to the ATR kinase may thus be sufficient to trigger phosphorylation.
To verify that the kinase pulled down by the Flag Ab was indeed ATR, we used two known ATR inhibitors, wortmannin and caffeine (1). ATM is more sensitive to wortmannin than is ATR, while the two kinases have similar sensitivities to caffeine. Our data indicate (Fig. 5C) that the kinase activity was clearly inhibited in a dose-dependent manner by wortmannin. Previous reports have shown that 1 μM wortmannin is sufficient to completely abolish ATM activity (1). As shown in Fig. 5C, we still detected a fair amount (∼50%) of kinase activity at that dose, suggesting that the kinase pulled down by the Flag Ab is indeed ATR. Because ATR and ATM substrate specificities overlap considerably (1), we also evaluated ATM activity on NPM. Our data indicate that NPM is also a good substrate for the Flag-ATM pulled down from RKO cells exposed to bleomycin (Fig. 5D, lane 4). ATR and ATM are both inhibited by caffeine, so this inhibitor was also used to verify the sensitivity of the pulled-down kinases. As shown in Fig. 5D, caffeine did indeed inhibit the activity of both kinases in a dose-dependent manner. To rule out the possibility that trace contamination of DNA-dependent protein kinase could be responsible for the kinase activity, we used DNase I in the reaction to digest a potential DNA component that could activate DNA-dependent protein kinase. Our data indicate (Fig. 5D) that DNase I is not affecting the kinase activity. Taken together, these data strongly suggest that NPM is a bona fide ATR and ATM substrate. To evaluate the possibility that NPM could compete with p53 for ATR, we compared the kinase activities on the two substrates. The data shown in Fig. 5E indicate that NPM is more avidly phosphorylated by ATR than is the recombinant GST-p53. The efficiency of ATR on NPM is even more compelling than the data show, since much less NPM than p53 protein was used in the assay (lanes 4 to 6). This could reflect a greater affinity of ATR for NPM or the presence of multiple phosphorylation sites.
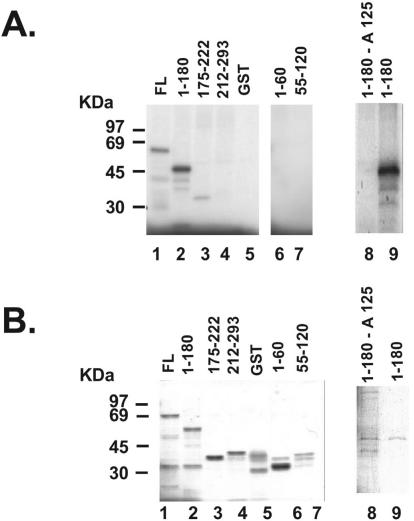
The consensus sequence for phosphorylation by ATM and its related kinase ATR overlaps extensively with the phosphatidylinositol kinase site. Generally the sequence Ser/Thr-Gln-Glu is targeted (1). Hydrophobic or acidic residues surrounding the Ser-Gln motif are favorable for phosphorylation, while positively charged amino acids are inhibitory. Even though no ATR-ATM consensus site was found on NPM primary sequence, the presence of several acidic residues (5) may increase the affinity of the kinases for the surrounding Ser. To determine the ATR phosphorylation site on NPM, we engineered several overlapping GST deletion mutants of NPM and used them in an in vitro ATR kinase assay. The data shown in Fig. 6A indicate that the most prominent Flag-ATR phosphorylation site is located between residues 1 and 180 (lane 2). Two additional overlapping mutants, 1 to 60 and 55 to 120, were then constructed to narrow down the phosphorylation site. Because none of the additional fragments were phosphorylated (lanes 6 and 7), we concluded that the phosphorylation site was located between residues 120 and 180. This fragment contains five Ser residues, located at positions 125, 137, 139, 143, and 149 (5). Almost 2 decades ago, Ser125 was identified as an in vivo NPM phosphorylation site (4). This residue was thus mutated to an alanine residue in the 1 to 180 fragment and assayed for phosphorylation by ATR. Our data (Fig. 6A, lanes 8 and 9) indicate that mutation of Ser125 to Ala completely abolished the phosphorylation of the fragment by ATR. These data indicate that Ser125 is the ATR phosphorylation site on NPM and that a single residue is responsible for the greater ATR activity on this protein than on GST-p53 (Fig. 5E). Ser125 has also been shown to be phosphorylated by casein kinase 2 (CK2) (4), but since CK2 is insensitive to wortmannin (8), we conclude that the Flag-ATR activity that we measured in Fig. 5 was indeed ATR and that ATR and CK2 share the same phosphorylation site on NPM.
FIG. 6.
Ser125 is the NPM ATR phosphorylation site. (A) NPM GST deletion mutants were constructed as described in Materials and Methods and phosphorylated by ATR in vitro. Mutation of Ser125 to alanine (lane 8) completely abolishes phosphorylation by ATR. (B) SDS-polyacrylamide gel electrophoresis of all the recombinant proteins used in the kinase assay (A).
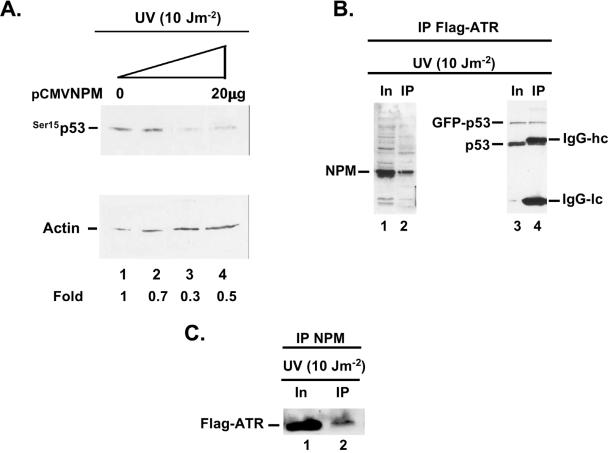
Because NPM is a substrate of ATR (Fig. 5) and seems to be preferentially phosphorylated by ATR (Fig. 5E), we then proceeded to determine whether NPM could compete with endogenous p53 for ATR phosphorylation. In order to do that, we transiently transfected increasing amounts of an NPM expression vector in human RKO colon carcinoma cells and then exposed the cells to 10 J of UV radiation m−2. As shown in Fig. 7A, low levels of p53 phosphorylation at Ser15 do occur at 10 J m−2 in the absence of NPM (lane 1). However, increasing amounts of NPM reduce considerably the levels of p53 phosphorylation at Ser15 (lanes 3 and 4). These data suggest that NPM could compete with p53 for ATR phosphorylation in vivo. Even though CK2 can also phosphorylate p53 in response to UV radiation (16), we believe that the inhibitory effect of NPM on p53 phosphorylation at Ser15 (Fig. 7A) is mediated through ATR competition because NPM binds to the p53 N terminus (Fig. 2) where ATR phosphorylates p53 at Ser15 and that the CK2 phosphorylation site on p53 is Ser392 at the C terminus (16).
FIG. 7.
(A) NPM reduces the levels of p53 phosphorylation at Ser15 by ATR in vivo. RKO cells were transiently transfected with increasing amounts of pCMV NPM, exposed to 10 J of UV radiation m−2 48 h after transfection, and harvested 4 h after UV radiation. Western blot analysis was performed on the cellular extracts with a p53 Ab specific for p53 phosphorylation at Ser15. Blots were reacted with actin Ab for a loading control. Fold activation of p53 Ser15 was measured by densitometry. (B) NPM and p53 form a complex in vivo with ATR. RKO cells stably transfected with a Flag-ATR expression vector were transiently transfected with a green fluorescent protein-p53 expression vector and exposed to 10 J of UV radiation m−2. The cellular extracts were immunoprecipitated with a Flag Ab and reacted with an NPM Ab (lanes 1 and 2) or a p53 Ab (lanes 3 and 4). Amounts of respective protein present in the input (In) and the immunoprecipitated fractions (IP) are indicated. IgG-hc, immunoglobulin heavy chains; IgG-lc, immunoglobulin light chains. (C) Reciprocal experiment with NPM Ab. RKO cells stably transfected with a Flag-ATR expression vector were exposed to 10 J of UV radiation m−2 and immunoprecipitated with an NPM Ab. The immunoprecipitated proteins were reacted with a Flag Ab. Abbreviations are as given for panel B.
In order to compete efficiently in vivo, it would be advantageous for NPM to form a complex with ATR and p53. To verify this concept, we stably transfected a Flag-ATR construct in RKO cells and then transiently transfected a GFP-p53 construct before exposing the cells to 10 J of UV radiation m−2. The advantage of this approach is that it facilitates immunoprecipitation of ATR and distinguishes GFP-p53 fusion from the endogenous p53, which migrates very closely to the immunoglobulin G (IgG) heavy chain. The data shown in Fig. 7B indicate that the Flag-ATR Ab immunoprecipitated efficiently the endogenous NPM (Fig. 7B, lane 2) as well as the GFP-p53 and the endogenous p53 protein (lane 4). In a reciprocal experiment, the NPM Ab could also immunoprecipitate ATR (Fig. 7C). These data support our concept that NPM forms a complex with ATR and p53 in vivo and could compete with p53 for ATR phosphorylation at low UV doses. Whether the three proteins exist together in a complex remains to be shown, but this possibility cannot be ruled out, since the NPM ATR site is located at Ser125 (Fig. 6) and the p53 interaction site is between residues 187 and 295 (7). NPM could thus bind to both ATR and p53 simultaneously.
The data presented here indicate that NPM is a natural repressor of p53 that may contribute to dampening p53 function during cellular growth or in the presence of low levels of DNA damage (28). To our knowledge, NPM is the first endogenous repressor reported to mediate this function. NPM may thus provide a threshold mechanism to sense the onset of genotoxic stress signals and prevent premature activation of p53. In addition, our data also suggest that overexpression of NPM could prevent normal p53 functions (Fig. 1). This possibility correlates well with NPM association with cell proliferation and tumor progression (5). An apparently contradictory report (7) indicated that NPM mediates stabilization of p53 and senescence (7). One possible explanation for this discrepancy may be the levels of NPM activity in the systems under study. For example, NPM is phosphorylated by CDK2/cyclin E (22, 27). Cyclin E expression is enhanced by low (10-J m−2) doses of UV radiation but inhibited by high (30-J m−2) doses of UV radiation (6). The study by Colombo et al. (7) used 25 J of UV radiation m−2, which may have been sufficient to inhibit cyclin E and NPM. This could explain why no induction of NPM by UV radiation was observed in the U2OS cell line that they studied.
The role of ATR in the UV response seems to be dose dependent. For example, doses as low as 1 J m−2 are sufficient to inhibit replication initiation and activate ATR (13). At higher doses, ATR is thought to prevent activation of late origins of replication and chromosome condensation. In contrast to ATM, ATR activity does not seem to increase in response to UV radiation but rather translocates into nuclear foci where it interacts with its substrates (1). Because ATR is active at very low doses of UV radiation, it thus seems essential for the cells to prevent its effect on p53, and possibly other substrates, to prevent premature cell cycle arrest and/or apoptosis. Our data indicate that NPM could mediate this function by setting a threshold for p53 activation by ATR (Fig. 3). Our data also imply that overexpression of NPM in cancer cells (5, 9) could contribute to p53 inactivation and cancer progression.
Acknowledgments
This work was supported by grant 1RO1GM57827-01 from the National Institutes of Health (to F.C.), a Graduate Program Enrichment Fellowship from the University of Maryland (D.A.M.), and an Initiative for Minority Student Development, GM55036 (D.A.M.).
We thank Steven Hirschfeld for careful reading of the manuscript and important discussions.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 3.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 4.Chan, P. K., M. Aldrich, R. G. Cook, and H. Busch. 1986. Amino acid sequence of protein B23 phosphorylation site. J. Biol. Chem. 261:1868-1872. [PubMed] [Google Scholar]
- 5.Chan, W. Y., Q. R. Liu, J. Borjigin, H. Busch, O. M. Rennert, L. A. Tease, and P. K. Chan. 1989. Characterization of the cDNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry 28:1033-1039. [DOI] [PubMed] [Google Scholar]
- 6.Chang, D., F. Chen, F. Zhang, B. C. McKay, and M. Ljungman. 1999. Dose-dependent effects of DNA-damaging agents on p53-mediated cell cycle arrest. Cell Growth Differ. 10:155-162. [PubMed] [Google Scholar]
- 7.Colombo, E., J. C. Marine, D. Danovi, B. Falini, and P. G. Pelicci. 2002. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 4:529-533. [DOI] [PubMed] [Google Scholar]
- 8.Davies, S. P., H. Reddy, M. Caivano, and P. Cohen. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derenzini, M. 2000. The AgNORs. Micron 31:117-120. [DOI] [PubMed] [Google Scholar]
- 10.Drexler, H. G., S. M. Gignac, R. von Wasielewski, M. Werner, and W. G. Dirks. 2000. Pathobiology of NPM-ALK and variant fusion genes in anaplastic large cell lymphoma and other lymphomas. Leukemia 14:1533-1559. [DOI] [PubMed] [Google Scholar]
- 11.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 12.Fornace, A. J., Jr., I. Alamo, Jr., and M. C. Hollander. 1988. DNA damage-inducible transcripts in mammalian cells. Proc. Natl. Acad. Sci. USA 85:8800-8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heffernan, T. P., D. A. Simpson, A. R. Frank, A. N. Heinloth, R. S. Paules, M. Cordeiro-Stone, and W. K. Kaufmann. 2002. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol. Cell. Biol. 22:8552-8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, P. S., and B. Y. Yung. 1999. Down-regulation of nucleophosmin/B23 mRNA delays the entry of cells into mitosis. Biochem. Biophys. Res. Commun. 257:865-870. [DOI] [PubMed] [Google Scholar]
- 15.Kapoor, M., R. Hamm, W. Yan, Y. Taya, and G. Lozano. 2000. Cooperative phosphorylation at multiple sites is required to activate p53 in response to UV radiation. Oncogene 19:358-364. [DOI] [PubMed] [Google Scholar]
- 16.Kapoor, M., and G. Lozano. 1998. Functional activation of p53 via phosphorylation following DNA damage by UV but not γ radiation. Proc. Natl. Acad. Sci. USA 95:2834-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kastan, M. B., Q. Zhan, W. S. el-Deiry, F. Carrier, T. Jacks, W. V. Walsh, B. S. Plunkett, B. Vogelstein, and A. J. Fornace, Jr. 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71:587-597. [DOI] [PubMed] [Google Scholar]
- 18.Kern, S. E., J. A. Pietenpol, S. Thiagalingam, A. Seymour, K. W. Kinzler, and B. Vogelstein. 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256:827-830. [DOI] [PubMed] [Google Scholar]
- 19.Kondo, T., N. Minamino, T. Nagamura-Inoue, M. Matsumoto, T. Taniguchi, and N. Tanaka. 1997. Identification and characterization of nucleophosmin/B23/numatrin which binds the anti-oncogenic transcription factor IRF-1 and manifests oncogenic activity. Oncogene 15:1275-1281. [DOI] [PubMed] [Google Scholar]
- 20.Lin, J., M. Blake, C. Tang, D. Zimmer, R. R. Rustandi, D. J. Weber, and F. Carrier. 2001. Inhibition of p53 transcriptional activity by the S100B calcium-binding protein. J. Biol. Chem. 276:35037-35041. [DOI] [PubMed] [Google Scholar]
- 21.Maeda, T., M. T. Chong, R. A. Espino, P. P. Chua, J. Q. Cao, E. G. Chomey, L. Luong, and V. A. Tron. 2002. Role of p21(Waf-1) in regulating the G1 and the G2/M checkpoints in ultraviolet-irradiated keratinocytes. J. Investig. Dermatol. 119:513-521. [DOI] [PubMed] [Google Scholar]
- 22.Okuda, M., H. F. Horn, P. Tarapore, Y. Tokuyama, A. G. Smulian, P. K. Chan, E. S. Knudsen, I. A. Hofmann, J. D. Snyder, K. E. Bove, and K. Fukasawa. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103:127-140. [DOI] [PubMed] [Google Scholar]
- 23.Parker, A., Y. Gu, W. Mahoney, S. H. Lee, K. K. Singh, and A. L. Lu. 2001. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J. Biol. Chem. 276:5547-5555. [DOI] [PubMed] [Google Scholar]
- 24.Siliciano, J. D., C. E. Canman, Y. Taya, K. Sakaguchi, E. Appella, and M. B. Kastan. 1997. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 11:3471-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takemura, M., K. Sato, M. Nishio, T. Akiyama, H. Umekawa, and S. Yoshida. 1999. Nucleolar protein B23.1 binds to retinoblastoma protein and synergistically stimulates DNA polymerase alpha activity. J. Biochem. (Tokyo) 125:904-909. [DOI] [PubMed] [Google Scholar]
- 26.Todd, M. D., M. J. Lee, J. L. Williams, J. M. Nalezny, P. Gee, M. B. Benjamin, and S. B. Farr. 1995. The CAT-Tox (L) assay: a sensitive and specific measure of stress-induced transcription in transformed human liver cells. Fundam. Appl. Toxicol. 28:118-128. [DOI] [PubMed] [Google Scholar]
- 27.Tokuyama, Y., H. F. Horn, K. Kawamura, P. Tarapore, and K. Fukasawa. 2001. Specific phosphorylation of nucleophosmin on Thr(199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J. Biol. Chem. 276:21529-21537. [DOI] [PubMed] [Google Scholar]
- 28.Vousden, K. H. 2002. Activation of the p53 tumor suppressor protein. Biochim. Biophys. Acta 1602:47-59. [DOI] [PubMed] [Google Scholar]
- 29.Wang, J. A., S. Fan, R. Q. Yuan, Y. X. Ma, Q. Meng, I. D. Goldberg, and E. M. Rosen. 1999. Ultraviolet radiation down-regulates expression of the cell-cycle inhibitor p21WAF1/CIP1 in human cancer cells independently of p53. Int. J. Radiat. Biol. 75:301-316. [DOI] [PubMed] [Google Scholar]
- 30.Wu, M. H., J. H. Chang, and B. Y. Yung. 2002. Resistance to UV-induced cell-killing in nucleophosmin/B23 over-expressed NIH 3T3 fibroblasts: enhancement of DNA repair and up-regulation of PCNA in association with nucleophosmin/B23 over-expression. Carcinogenesis 23:93-100. [DOI] [PubMed] [Google Scholar]
- 31.Yang, C., D. A. Maiguel, and F. Carrier. 2002. Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res. 30:2251-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhan, Q., F. Carrier, and A. J. Fornace, Jr. 1993. Induction of cellular p53 activity by DNA-damaging agents and growth arrest. Mol. Cell. Biol. 13:4242-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]