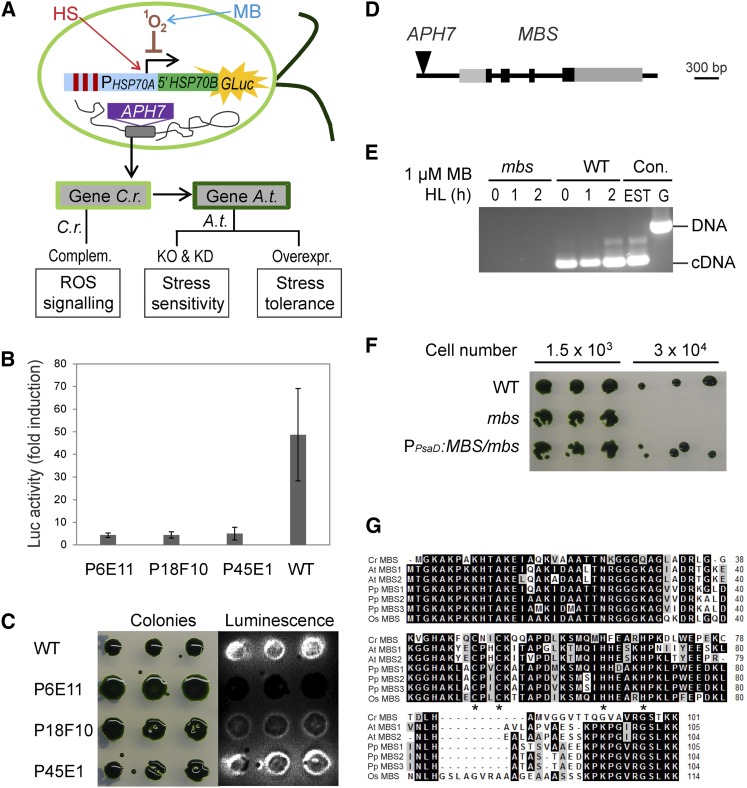
Figure 1.
Screen for ROS Signaling Mutants in C. reinhardtii and Identification of the MBS Gene.
(A) Schematic overview of the experimental strategy for isolating regulators of ROS signaling. High-throughput genetic screens with a ROS-inducible reporter gene in C. reinhardtii are combined with reverse genetic analyses in Arabidopsis. An algal strain expressing a 1O2-inducible luciferase reporter gene (PHSP70A-5′HSP70B-GLuc; Shao and Bock, 2008) was subjected to insertional mutagenesis with the hygromycin resistance gene APH7 (Berthold et al., 2002). Screening for mutants defective in 1O2-mediated signaling was performed by treatment of cultures with the photosensitizer MB that generates 1O2 in the light. As the HSP70A promoter also responds to heat stress (HS; heat stress elements indicated as red bars), ROS signaling mutants can be identified as strains displaying luciferase induction upon heat stress, but lacking induction upon exposure to 1O2 stress. Assays performed on mutants and transgenic lines are indicated in the boxes at the bottom of the panel. C.r., C. reinhardtii; A.t., Arabidopsis; Complem., complementation; KO, knockout; KD, knockdown; Overexpr., overexpression.
(B) Example of three insertion mutants (P6E11, P18F10, and P45E1) that do not respond to 1O2 stress generated by HL (1000 µE m−2 s−1) in the presence of 1 µM MB (monitored by luciferase bioluminescence). Data are mean values ± sd of three biological replicates. WT, wild type.
(C) Heat stress response in the three insertion mutants P6E11, P18F10, and P45E1. Growing algal colonies were shifted from 23 to 40°C for 1 h. While the P45E1 mutant has a normal heat stress response (as evidenced by induction of luciferase luminescence), the P6E11 and P18F10 mutants show an impaired response to heat stress compared with the wild type.
(D) Model of the MBS gene and insertion site of the APH7 marker in the mbs mutant of C. reinhardtii. Black boxes represent exons, and gray boxes represent 5′ and 3′ untranslated regions as confirmed by EST sequencing.
(E) RT-PCR analysis demonstrating the absence of MBS transcripts in mutant P45E1 (mbs). The 300-bp MBS gene-specific RT-PCR product is obtained with wild-type cDNA as template after 0, 1, and 2 h of HL treatment (1000 µE m−2 s−1) in the presence of 1 µM MB but cannot be amplified from cDNA of the mbs mutant. PCR products amplified from an EST clone (AV621044) and genomic DNA (G) of the wild type are shown as controls (Con.).
(F) MB-sensitive phenotype of the mbs mutant (P45E1) compared with the wild type and the complemented PPsaD:MBS/mbs strain.
(G) Alignment of the amino acid sequences of the MBS proteins from the alga C. reinhardtii (Cr), the moss P. patens (Pp), and the higher plants Arabidopsis (At) and rice (Os). Putative MBS sequences were retrieved from the full genome sequences and the alignment was produced using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The asterisks indicate the zinc finger motif of the C2H2 type. Amino acids highlighted in black indicate identical residues in all sequences, and residues in gray denote similar amino acids.