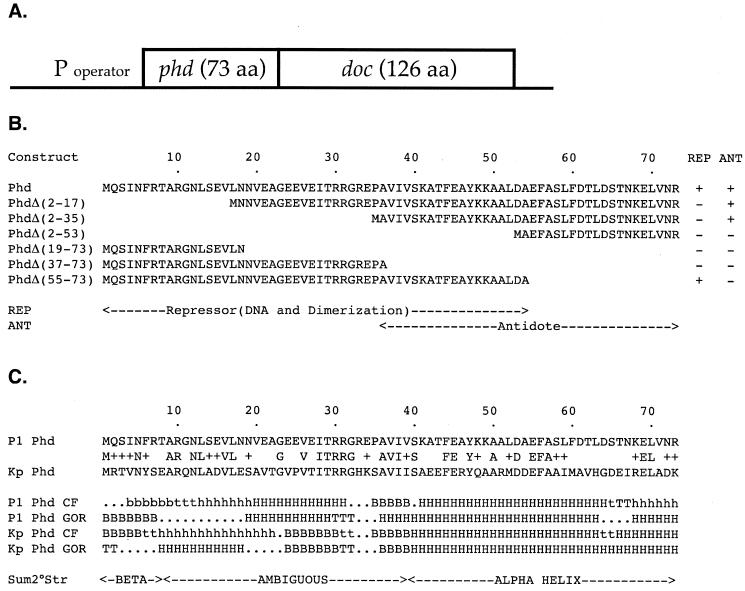
FIG. 1.
(A) Schematic structure of the P1 addiction operon. (B) Deletion analysis of Phd: predicted protein products and summary of their activities. The predicted protein products of phd and various deletion mutants of phd are shown. The repressor (REP) and antitoxin (ANT) activities of each construct are indicated. Repressor activity was indicated by the ability to repress transcription of a lacZ reporter fused to the P1 addiction promoter (Table 3). Antitoxin activity was indicated by the ability of the cells to grow in the presence of an otherwise lethal level of the Doc toxin (Table 4). (C) Secondary structure predictions for Phd and a Klebsiella homolog. The aligned sequences of Phd and a Phd homolog from K. pneumoniae (Kp) are given along with their predicted secondary structures, as determined by the Chou-Fasman algorithm (CF) (8) and by the Garnier-Osguthorpe-Robson algorithm (GOR) (16).