Abstract
OP9 is a yet-uncultivated bacterial lineage found in geothermal systems, petroleum reservoirs, anaerobic digesters, and wastewater treatment facilities. Here we use single-cell and metagenome sequencing to obtain two distinct, nearly-complete OP9 genomes, one constructed from single cells sorted from hot spring sediments and the other derived from binned metagenomic contigs from an in situ-enriched cellulolytic, thermophilic community. Phylogenomic analyses support the designation of OP9 as a candidate phylum for which we propose the name ‘Atribacteria’. Although a plurality of predicted proteins is most similar to those from Firmicutes, the presence of key genes suggests a diderm cell envelope. Metabolic reconstruction from the core genome suggests an anaerobic lifestyle based on sugar fermentation by Embden-Meyerhof glycolysis with production of hydrogen, acetate, and ethanol. Putative glycohydrolases and an endoglucanase may enable catabolism of (hemi)cellulose in thermal environments. This study lays a foundation for understanding the physiology and ecological role of the ‘Atribacteria’.
Introduction
Over the last ~20 years, cultivation-independent approaches in microbial ecology have dramatically expanded our view of the microbial world1,2 and have revealed that our ability to isolate novel organisms out of the milieu for study remains limited. While this problem is evident at all taxonomic levels, it is most glaring at the phylum level. Currently, <50% of phylum-level lineages of Bacteria and Archaea have been cultivated and studied in the laboratory2,3. The vast diversity of these uncultured groups represents an enormous genetic reservoir that has been described as ‘biological dark matter’ to call attention to our profound ignorance of these groups4. It can be difficult to discover even the most basic facts about the biology of candidate microbial phyla because they are often outnumbered in nature.
Metagenomics and single-cell genomics are powerful cultivation-independent approaches for probing the nature of so-called ‘dark matter’ organisms by facilitating access to their genomes5,6. While metagenomics has allowed the recovery of partial or nearly-complete genomes from several candidate phyla in habitats where they are naturally abundant7–9, access to less abundant ‘dark matter’ groups has been enhanced by the advent of single-cell genomics, where individual cells are isolated by fluorescence activated cell sorting (FACS) or micromanipulation techniques including microfluidic sorting, optical trapping, and micropipetting; isolated cells are then lysed and the femtogram quantities of DNA released are amplified and sequenced5,6. Through containment of the sorting and amplification steps within nanoliter reaction volumes, microfluidic approaches ameliorate amplification bias, minimize the amplification of trace DNA contaminants in the sample, laboratory environment, and reagents, and allow for detailed observation of cell morphology10–12. Complete or partial genomes can be obtained from single cells10,13,14 and single-cell genomics has shed light on the possible functions of several candidate phyla4,15–18.
Here, we take advantage of the complementarity of metagenomics and single-cell genomics to assemble nearly-complete genomes of two members of candidate bacterial phylum OP9. Since its discovery in Obsidian Pool in Yellowstone National Park (YNP)1, OP9 has been found in other geothermal springs, petroleum reservoirs, thermal bioreactors and digesters, and wastewater sludge treatment plants19–23. This distribution demonstrates an affinity for thermal, anaerobic environments, but no genetic data pertaining to OP9 other than PCR-amplified 16S rRNA gene sequences have been reported. We compare single-cell genomic data from 15 OP9 cells isolated from hot spring sediments near Little Hot Creek, CA, (LHC)19 with a metagenomic dataset from an in situ, ~77 °C cellulolytic enrichment in Great Boiling Spring, NV (GBS)20 to obtain distinct draft genomes of the OP9 population from each environment. Analysis of the shared features of these two genomes offers the first insights into the cell structure and metabolic capabilities of OP9 lineages present in geothermal systems and suggests that they play a role in biomass degradation in these environments.
Results
Morphology-based single-cell sorting targeting OP9
Single-cell sorting, lysis, and whole genome amplification using an optical trap and microfluidic device14,17, as described in Methods, was performed on cells separated from ~80 °C sediments of the hot spring LHC4 (Supplementary Figure S1), which were previously shown to be dominated by novel Archaea and Bacteria19. Initial efforts surveyed the morphological diversity, with the aim of identifying morphologically distinct ‘dark matter’ groups that could be targeted in subsequent sorting efforts. Rod-shaped cells (average 0.5 μm × 4.5 μm) were identified as members of the candidate phylum OP9. This morphology was distinctive in the sample and was targeted in two subsequent sorting efforts, the second of which resulted in 15 of 21 OP9 single cell genomes (SCGs), as assessed by PCR screens. Thus, the morphology-based sorting approach was effective for this lineage and sample, even though it was present at a relatively low abundance (~0.5% of total cells). Half of the OP9 SCGs obtained had 16S rRNA gene fragments identical to the OP9 sequence previously described from LHC4 (LHC4_L1_A09)19, while the others differed by 1-2 bases over the ~600 bp sequenced region. This indicated that the sorted OP9 cells all represented a closely related group and that this lineage was a stable member of the microbial community in LHC4 sediments, given that OP9 was recovered from samples collected on three dates spanning ~22 months.
Assembly of a composite OP9 single cell genome
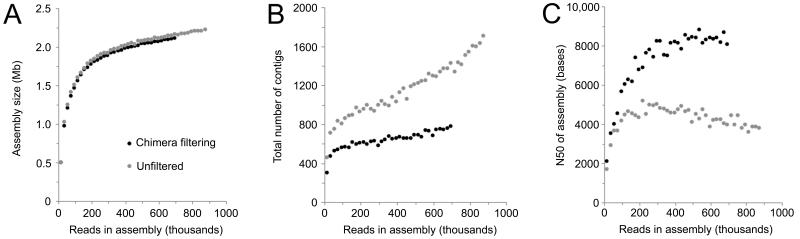
After pyrosequencing, filtering, and assembly as described in Methods, 15 SCGs with uniform read %G+C content and contig tetranucleotide word frequencies (TNF) were chosen for further sequencing and reassembly (Supplementary Figure S2). Resulting individual de novo SCG assemblies ranged from 110-872 kb (Supplementary Table S1). The average nucleotide identity (ANI) between overlapping regions of the SCGs ranged from 96.2-99.4% with an average of 98.8% per SCG, greater than the genomic 95% ANI empirically determined to delineate species24. A neighbor-joining tree using a distance matrix based on pairwise %ANI did not reveal distinct clusters within the SCGs (Supplementary Figure S3). Along with high 16S rRNA gene identity (>99.6%), these data indicate that the 15 SCGs represent a single, species-level group within the OP9 lineage. Given the small initial de novo assembly sizes and the relatively low sequencing depth for SCGs10 (1.5-20x, assuming a 1.5-3.0 Mb genome typical of thermophiles), we determined that it would be difficult to assemble nearly-complete single-cell genomes from these datasets individually. However, because the 15 OP9 SCGs represented a single species, we combined the datasets to construct a composite SCG (cSCG) assembly. Pooling data from multiple cells alleviates problems inherent in single-cell genomics, such as the random bias in genomic coverage of individual SCGs arising from whole genome amplification by multiple strand-displacement amplification (MDA)10, and resulted in an assembly of ~2.24 Mb (Table 1). Integration of the multiple SCG datasets also enabled application of a jackknifing procedure for removal of chimeric sequences14, which occur at a frequency of one chimera per ~10 kb during MDA10. This reference-independent chimera filtering procedure significantly improved the assembly, as evidenced by a >2-fold increase in contig N50 with only a small decrease in overall assembly size despite removal of ~18.5% of total reads (Figure 1, Table 1).
Table 1.
Assembly statistics for the OP9-cSCG and OP9-77CS
| Assembly | # of reads |
Assembly size, total # of bases |
# of contigs (scaffolds)c |
Contig (or scaffold)c N50, bases |
Largest contig (or scaffold)c, bases |
|---|---|---|---|---|---|
| Unfiltered OP9-cSCG | 862139 | 2241199 | 1718 | 3838 | 27213 |
| Chimera-filtered OP9-cSCG | 702720 | 2124461 | 780 | 8210 | 38188 |
| 545 | 15714 | 78011 | |||
| Scaffolded OP9-cSCGa | n.d.b | 2098790 | |||
| (390) | (63552) | (216971) | |||
| OP9-77CS metagenome bina | n.d.b | 2252356 | 306 | 14273 | 59788 |
After removal of potentially contaminating contigs as described in Results; these filtered assemblies were used for analyses.
Not determined
Numbers in parentheses refer to the number or sizes of scaffolds in the scaffolded assembly.
Figure 1.
Chimera-filtering increases contig N50 in the cSCG assembly. Stepwise assembly of composite OP9 SCG before (grey points) or after (black points) filtering out potentially chimeric reads. The increase in (A) total assembly size, (B) number of contigs, and (C) contig N50 are shown as a function of the number of reads (sampling with replacement) used.
Identification of OP9 contigs in a cellulolytic metagenome
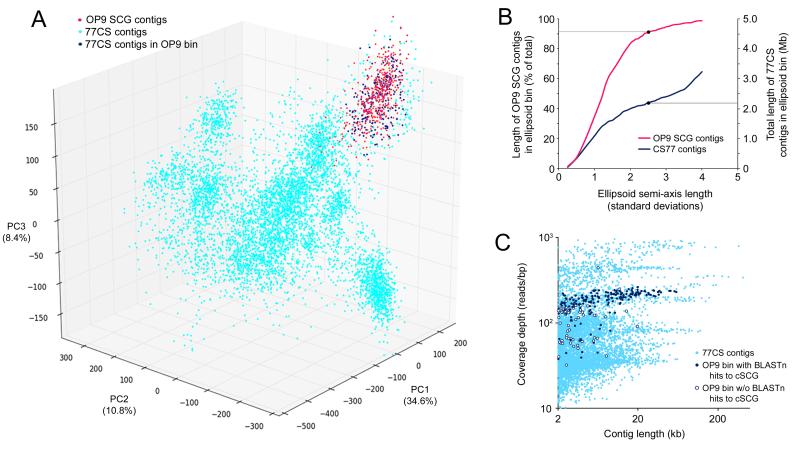
Principal components analysis of TNF (TNF-PCA) was used to probe metagenomes derived from LHC and another hot spring in the US Great Basin, GBS18, located ~400 km north of LHC. Although OP9 reads were present in extremely low abundance in the LHC metagenome, pyrosequencing of 16S rRNA gene fragments amplified from three cellulolytic enrichments incubated at ~77 °C in GBS indicated significant enrichment of OP9 in comparison to undisturbed GBS sediments25. TNF-PCA revealed a distinct cluster of contigs from one of these metagenomes (corn stover incubated in spring sediment, ‘77CS’) overlapping with OP9 SCG contigs (Figure 2A), suggesting that this cluster represented genomic fragments from the enriched OP9 lineage, and allowed designation of an OP9 bin (‘OP9-77CS’; Figure 2A, B) of 315 contigs containing ~2.23 Mb. The majority of OP9-77CS contigs displayed a high level of identity to the OP9-cSCG by BLASTN and exhibited a defined read depth within the metagenome (213 ± 50 s.d.; Figure 2C), suggesting a single OP9 phylotype closely related to the OP9-cSCG. While a portion of OP9-77CS contigs had lower read depth, ~40% of these had >85% identity to the OP9-cSCG, and may be enriched for genes unique to an OP9 strain present at lower abundance in the enrichment. Although some of the contigs in the OP9-77CS bin without BLASTN hits to the cSCG may have originated from organisms other than OP9, they could also represent portions of the OP9 genome that were either not covered or not present in the OP9-cSCG, and were thus retained.
Figure 2.
Identification of OP9-like contigs from the 77CS metagenome. (A) Principal component analysis (PCA) of tetranucleotide frequency of contigs in the 77CS metagenome and ten individual OP9 SCG assemblies. 77CS contigs in dark blue were binned as OP9-like, as defined by their placement within an ellipsoid with a centroid and semi-axis lengths equal to the mean and 2.5 times the standard deviation, respectively, of the first three principle components (PC1-PC3, with the percent variation explained by each in parentheses) of the OP9 SCG contigs. Only contigs greater than 2 kb in length are shown. Contigs <2kb inside this ellipsoid were also included in the OP9 bin if they had significant nucleotide identity (>85% identity over >100 nt) to contigs in the OP9-cSCG by BLASTn. (B) Total length of OP9 SCG and 77CS contigs >2kb contained within ellipsoids with different semi-axis lengths defined by multiplication of the standard deviation of PC1-PC3 of the OP9 SCG contigs by increasing scalar quantities (x-axis). Black points on the curves indicate the multiplier (2.5) used to define the OP9 bin, chosen at an approximate inflection point of the CS77 contig length curve to maximize inclusion of metagenome contigs in the cluster overlapping the OP9 SCGs but minimize inclusion of contigs in adjacent clusters. (C) Plot of coverage depth vs. contig length for the CS77 metagenome assembly, highlighting contigs in the OP9 bin with (dark blue circles) and without (open circles) >85% nucleotide identity over >100 bp to the OP9-cSCG assembly.
Comparison and estimation of completeness of the OP9 genomes
The OP9-cSCG and OP9-77CS assemblies displayed a high level of identity and likely represent distinct but closely related species. Each contained full-length 16S rRNA genes that were 98.6% identical. BLASTN revealed reciprocal cSCG-to-77CS coverage of 87.7% and 81.6% with an ANI of 91.9%. These were below the accepted thresholds for 16S rRNA gene identity (98.7%) and genomic ANI (95%) delineating microbial species3; thus, co-assembly of the cSCG and OP9-77CS genomes was not justified. However, the homology and high coverage between the two datasets allowed us to use the OP9-77CS contigs as scaffolds for the cSCG. This scaffolding identified short overlaps among existing contigs and improved the cSCG assembly significantly (Table 1). Because of the high degree of coverage and identity between the two datasets, cross-comparison allowed a rigorous filtering of potential contaminating sequences that could not be accomplished with either dataset in isolation. Such filtering, described in Methods, resulted in removal of 9 and 13 contigs from OP9-77CS and OP9-cSCG, respectively, representing ~1% of the assemblies. We have conservatively restricted our prediction of structure and function to genes and pathways present in both the OP9-cSCG and OP9-77CS, representing the core genome from the OP9 lineage present in Great Basin hot springs.
Analysis of the predicted protein-encoding genes (coding DNA sequences, CDSs) in the OP9-cSCG and OP9-77CS supported their close relationship and their novelty compared to other genomes (Table 2). 1760 CDSs were identified as best-bidirectional hits between the two genomes, with an average predicted protein sequence identity of 94.3%; most CDSs with no BLASTP hit in the other OP9 genome were annotated as hypothetical proteins (73% and 82% for comparison of the 77CS-to-cSCG and cSCG-to-77CS, respectively). The number of CDSs assigned EC numbers by RAST, KAAS and Markov clustering was similar (22.8-35%). The percentages of OP9 CDSs assigned to Clusters of Orthologous Groups (COGs)26 were 64.6-68%, notably lower than the average of ~80% for genomes of cultivated microbes27 but comparable to other genomes from previously unsequenced phyla, which are typically enriched with novel CDSs17.
Table 2.
Genomic features of OP9-cSCG and OP9-77CS
| OP9-cSCG | OP9-77CS | |
|---|---|---|
| G+C content | 55.4% | 55.8% |
| Number of predicted CDSs | 2289 | 2478 |
| PEGs with predicted function | ||
| Non-“hypothetical” (RAST) | 1545 | 1789 |
| COG assigned (CDD) | 1557 | 1601 |
| Pfam(s) assigned (CDD) | 1423 | 1581 |
| KO assigned (KAAS) | 1068 | 1161 |
| EC # assigned (RAST) | 523 | 585 |
| EC # assigned (KAAS) | 545 | 601 |
| EC # assigned (Markov) | 748 | 858 |
| No. of RNAs | ||
| tRNAs | 44 | 57 |
| 23S rRNAs | 1 | 3b |
| 16S rRNAs | 1 | 1 |
| 5S rRNAs | 1 | 1 |
| Estimated completenessa | 96.4% | 98.2% |
Based on presence of conserved markers exclusive of those involved in de novo riboflavin and pantothenate synthesis, see text.
One full length 23S rRNA gene was observed, in addition to two partial sequences present on small contigs; all three were >99% identical over their entire length to the corresponding gene in the OP9-cSCG.
The presence of essential features and highly conserved genes in the OP9-cSCG and OP9-77CS suggested that both datasets represent nearly-complete genomes (Table 2). Each contains a complete set of aminoacyl-tRNA synthetases, at least one tRNA gene for each amino acid, and full-length 5S, 16S and 23S rRNA genes. Additionally, all 31 highly conserved genes recognized by AMPHORA28 were present. An approximate quantification of genome completeness was obtained by comparison to a set of 181 conserved, typically unlinked genes29; the OP9-cSCG and OP9-77CS contained 164 (90.6%) and 167 (92.3%) of these markers, respectively (Supplementary Table S2). However, a majority of the absent markers were genes involved in synthesis of pantothenate and riboflavin, precursors of coenzyme-A (Co-A) and flavin adenine dinucleotide (FAD), respectively. The presence of energy-coupling factor (ECF) transporters specific for these precursors, as well as all necessary genes to subsequently convert them into Co-A and FAD, suggests that the absence of genes for synthesis of pantothenate and riboflavin is due to dependence of OP9 on uptake of these substrates. If these 13 markers are excluded from the analysis, estimated completeness of the cSCG and OP9-77CS are >96% (Table 2), providing a level of confidence for inference of the presence and absence of metabolic capabilities based on gene content. Of the markers detected, only a small minority were present in greater than one copy in each assembly. In approximately half of these cases the extra markers were present on large contigs and were conserved (>90% identical and in the same genomic context) in both OP9 datasets, suggesting that they are genuinely present in multiple copies. When these are excluded, only 1.8% (3/164) and 4.8% (8/167) of the observed markers were present in greater than one copy. Given that not all of the markers are present in a single copy in all organisms, this analysis suggests a high fidelity for the assemblies, particularly for the OP9-cSCG.
Phylogeny and cell envelope structure
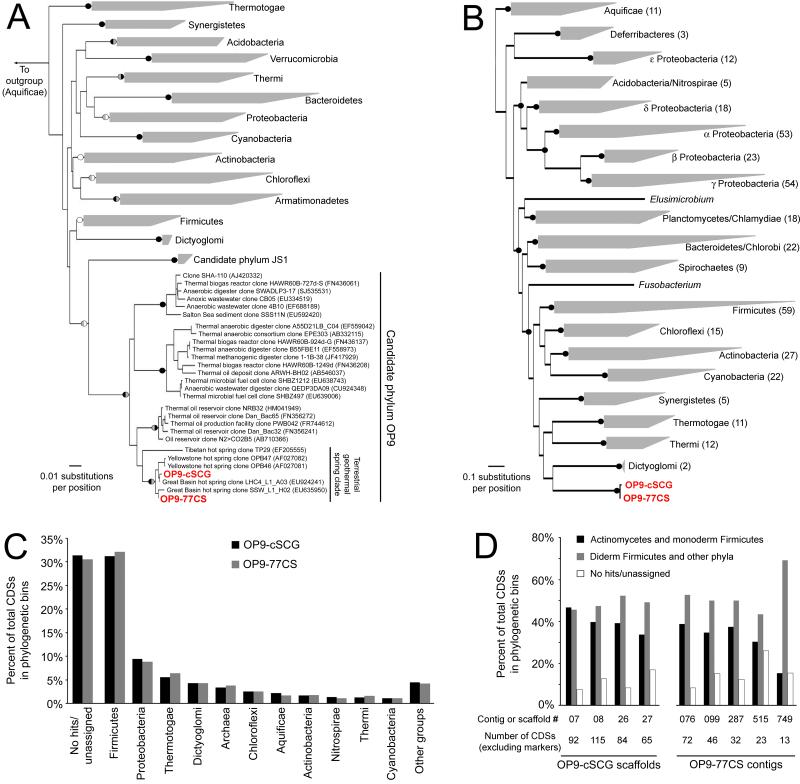
Phylogenies inferred from 16S rRNA gene sequences (Figure 3A) and 31 conserved protein-coding genes (Figure 3B) support the designation of OP9 as a phylum-level lineage in the Bacteria. OP9 is comprised of four distinct clades in the 16S rRNA gene-based phylogeny. The OP9-cSCG and OP9-77CS fall within one family-level clade, which inhabits terrestrial geothermal environments world-wide, including the US Great Basin, YNP, China, and Kamchatka (Figure 3A; additional OP9 sequences and references in Supplementary Table S3 footnote). Although the OP9 lineage clustered with the JS1 clade with weak bootstrap support, JS1 has been defined as a candidate phylum in its own right by some analyses30. Rigorous determination of whether these two clades represent distinct phyla or classes within a single phylum will require analysis of additional phylogenetic markers from JS1, which are currently unavailable.
Figure 3.
Relationship of OP9-SCG and OP9-CS77 to other bacterial groups. (A) Neighbor-joining tree based on a distance matrix of 1349 aligned positions of 16S rRNA genes from selected bacterial phyla with cultured representatives and the candidate phyla JS1 and OP9, including those in the OP9-SCG and OP9-CS77 assemblies. Black (100%), grey (>80%), and white (>50%) circles indicate bootstrap support (100 pseudoreplicates) for selected nodes in phylogenies inferred using neighbor-joining (left half of circle) and maximum-likelihood (right half) methods. (B) Maximum-likelihood phylogeny inferred from concatenated alignments of predicted amino acid sequences of 31 housekeeping genes identified by AMPHROA in genomes representing a variety of bacterial phyla and the OP9-SCG and OP9-CS77 assemblies. The number of genomes represented in each wedge is indicated in parentheses, and black circles indicate bootstrap support of >80% for 100 pseudoreplicates. (C) Binning of CDSs in the OP9 assemblies based on the phylogenetic affiliation of their top BLASTP hit to a database of sequenced bacterial and archaeal genomes. Phylogenetic groups with fewer than 1% of top hits were aggregated (‘other groups’). (D) Phylogenetic binning of CDSs on contigs or scaffolds containing markers diagnostic for a diderm cell envelope structure31, emphasizing that top BLASTP hits were distributed among both monoderm and diderm Bacteria. Diderm Firmicutes includes members of the Negativicutes and Halanaerobiales.
Genome-wide comparison of the OP9-cSCG and OP9-77CS with other Bacteria suggested a relationship with Gram-positive members of the phylum Firmicutes, although key genes indicated that OP9 is diderm (i.e. has an outer membrane). A plurality of predicted proteins in the OP9 genomes had top BLASTP hits to members of the Firmicutes, both when all CDSs were considered (Figure 3C) and when CDSs assigned to COGs were first divided into 20 major functional COG categories, ranging from 13 to 244 CDSs for a given category. The majority (>85%) of these top BLASTP hits were to proteins in members of the Firmicutes known or expected to have a monoderm, Gram-positive-like cell envelope structure. However, several genetic markers found exclusively in diderm Bacteria were present in both OP9 genomes, including proteins involved in outer membrane protein assembly (BamA/YaeT, OmpH), secretion across the outer membrane (TolC, TonB, secretin), and the flagellar P- and L-rings (along with other flagellar and chemotaxis genes) associated with the peptidoglycan and outer membrane in diderm Bacteria (Supplementary Table S4)31. Top BLASTP hits (average of 48% identity) of CDSs in contigs or scaffolds containing these markers were distributed in both monoderm and diderm taxa (Figure 3D) and were generally similar to the distribution of top BLASTP hits of CDSs throughout the assemblies, suggesting that these markers are genuine parts of the OP9-cSCG and OP9-77CS lineages and not recently obtained by horizontal gene transfer from known diderm taxa. Genes involved in synthesis and export of lipid A (lpxABCD; msbA), a core component of the outer membrane of many diderm Bacteria, were present, but no recognizable homologs of genes required for production and attachment of keto-deoxyoctulosonate and the liposaccharide inner core were found. The OP9 outer membrane structure therefore likely differs from that of other diderm Bacteria containing typical lipopolysaccharide.
Potential role of OP9 in (hemi)cellulolysis
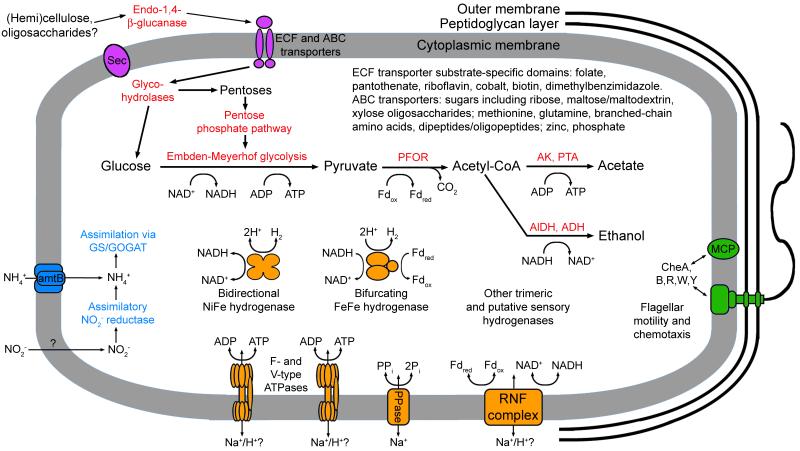
The presence of CDSs for both extracellular and cytoplasmic glycohydrolases indicate the potential for degradation and utilization of cellulose or hemicellulose by OP9. Both genomes encode an endo-1,4-β-glucanase with a putative N-terminal secretion signal peptide, indicating potential for extracellular activity (Figure 4). The closest characterized homologs of this gene (31-38% amino acid identity) are an endoglucanase in Pyrococcus furiosus (EglA)32 and a cellulase in Thermotoga maritima (CelA)33, both of which belong to the GH12 family of glycohydrolases34. Alignment of the OP9 endo-1,4-β-glucanases with members of GH12 indicate that two glutamates predicted to be involved in catalysis are conserved in the OP9 CDSs (positions 167 and 256). The T. maritima CelA is active on soluble glucans, while the P. furiosus EglA is active on crystalline cellulose and glucan oligosaccharides; both enzymes displayed weak activity on xylan32,33. Several members of the GH12 family have the capacity for hydrolysis of xyloglucan, a major component of hemicellulose in primary plant cell walls, and it has been suggested that xyloglucanase activity may be an ancestral trait of the GH12 family35. While it is difficult to predict the precise activities of the OP9 endo-1,4-β-glucanase, it is likely involved in hydrolysis of one or more components of (hemi)cellulose. OP9 apparently lacks the capacity for degradation of the lignin component in lignocellulose, as only a single putative dioxygenase (COG1355) and no feruloyl esterases (e.g. pfam07519) were observed in the assemblies.
Figure 4.
Overview of features and potential metabolic capabilities of the OP9 lineage represented by the OP9-SCG and OP9-CS77 genomes as discussed in the text. CDSs in the OP9-cSCG and OP9-77CS associated with predicted functions are listed in Supplementary Table S5. Proteins involved in specific processes are identified by color: secretion and transporters (purple); saccharide catabolism and fermentation (red); energy conservation (orange); flagellar motility and chemotaxis (green); and nitrogen transport and assimilation (blue). Substrates and products are not necessarily balanced in the reactions depicted. Abbreviations not indicated in the text: ABC, ATP-binding cassette; Fdox, oxidized ferredoxin; AK, acetate kinase; PTA, phosphotransacetylase; AlDH, aldehyde dehydrogenase; ADH, alcohol dehydrogenase; PPi, pyrophosphate; Pi, inorganic phosphate.
Several cytoplasmic glycohydrolases encoded in the OP9 genomes could allow for utilization of oligosaccharides derived from cellulose and hemicelluloses degradation, including xyloglucan. Both OP9 genomes encode an α-xylanase and β-glucosidase for degradation of isoprimeverose subunits of xyloglucan into glucose and xylose36, as well as an α-N-arabinofuranosidase that could act on the L-arabinose-substituted isoprimeverose. The OP9-77CS additionally encodes a β-galactosidase and α-L-fucosidase, which would allow for complete breakdown of all major types of xyloglucan oligomers into hexoses and pentoses. The β-glucosidase would also mediate hydrolysis of cellobiose and longer chain β-glucans. Both genomes additionally encode a β-xylanase, β-mannosidase, and amylopullulanase that could facilitate hydrolysis of xylan, mannan, and alpha-linked glucan oligosaccharides, respectively.
Central metabolism
Catabolic capacity of the OP9-cSCG and OP9-77CS appears mainly limited to utilization of hexoses and pentoses via Embden-Meyerhof glycolysis and the pentose phosphate pathway (Figure 4). The resulting pyruvate could be converted by pyruvate-ferredoxin oxidoreductase (PFOR) to acetyl-coenzyme A (acetyl-CoA), which could either be processed to acetate with concomitant production of ATP via acetate kinase and phosphoacetyl transferase or be reduced to ethanol by aldehyde and alcohol dehydrogenases. An incomplete tricarboxylic acid cycle and the absence of most genes for NADH:ubiquinone oxidoreductase (Respiratory Complex 1), cytochromes, cytochrome oxidases, and quinone synthesis suggests a reliance on fermentation, while the absence of catalase and presence of oxygen-sensitive enzymes such as PFOR indicate an anaerobic lifestyle. Ethanol and acetate, along with carbon dioxide and H2 (see below) are likely the major fermentation products, as genes involved in production of lactate and butyrate and other common fermentation pathways were not detected. Pathways for catabolism of amino acids and fatty acids are largely absent. While prediction of the full metabolic capacity is difficult due to the novelty of OP9, these data nonetheless suggest a prominent role for saccharide fermentation and are consistent the enrichment of OP9 on (hemi)cellulose substrates in GBS.
The OP9 genomes encode anabolic pathways for most important metabolites, but appear to be dependent on uptake of several vitamins. Neither genome contained evidence for carbon fixation pathways, indicating a heterotrophic lifestyle. Complete or nearly complete biosynthetic pathways for all amino acids except methionine were detected, as well as apparent capacity for de novo synthesis of purines, pyrimidines, NAD(P), and fatty acids. While OP9 has the capacity for synthesis of pyridoxine, it lacks pathways for synthesis of most other vitamins. The uptake of biotin, folate, pantothenate, riboflavin, and dimethylbenzimidazole is likely mediated by ECF transporters, as substrate binding domains specific for these vitamins are all present37. Ammonia uptake and assimilation appears to be mediated by an Amt transporter and the glutamine synthetase/glutamate-2-oxoglutarate amino transferase (GS/GOGAT) pathway, respectively. Two CDSs in the GS/GOGAT locus with similarity to assimilatory nitrite reductases may allow utilization of nitrite as an alternative nitrogen source, but no transporters for nitrite were detected. OP9 appears to be dependent on sulfide as a sulfur source because genes encoding assimilatory sulfate reductase were not present.
Energy conservation
In addition to ethanol production, regeneration of oxidized electron carriers could be mediated by several cytoplasmic hydrogenases, including one NiFe hydrogenase and four FeFe hydrogenases (Figure 4). The large subunit of the tetrameric NiFe hydrogenase clusters with group 3b bifunctional hydrogenases (Supplementary Figure S4)38, which includes sulfhydrogenases of thermophilic Archaea that can catalyze reduction of sulfur to sulfide in addition to hydrogen production39. Two of the FeFe hydrogenases cluster with group A1 and A8 trimeric NAD(P)-linked hydrogenases (Supplementary Figure S5)40. The subunits of one of these shares 44%-56% identity to those of a recently described ‘bifurcating’ hydrogenase in Thermotoga maritima, which couples H2 production to concomitant oxidation of NADH and reduced ferredoxin (Fdred)41 and may allow for proton reduction at relatively high H2 concentrations. The remaining two FeFe hydrogenases cluster with groups C and D, which are poorly characterized but may have a regulatory role due to the presence of PAS domains (Group C) and their association with protein kinases and phosphatases40.
Maintenance of a chemiosmotic membrane potential appears to be mediated primarily by an RNF complex, a membrane-associated pyrophosphatase (PPase), and two ATP synthases. RNF is a membrane-bound, six-subunit complex capable of energy conservation by coupling ferredoxin:NAD+ oxidoreductase activity with H+ or Na+ transport42. Depending on the intracellular redox state (balance of NADH and Fdred), the RNF complex could either allow production of a membrane potential by oxidation of NAD+ with Fdred, or production of Fdred from NADH by reverse electron transport. Accordingly, two ATP synthases (F-type and V-type) could allow for either ATP production from a chemiosmotic membrane gradient generated by the RNF complex or generation of membrane potential by ATP hydrolysis. The PPase and F-type ATP synthase appear to mediate Na+ and H+ transport, respectively, based on several conserved residues (Supplementary Figure S6)43,44, suggesting that both Na+ and H+ are involved in maintenance of membrane potential.
Discussion
The OP9-cSCG and OP9-77CS genomes described here offer the first significant insights into the metabolic capabilities and ecology of this ‘dark matter’ group. Our analysis of the core genome of hot spring OP9 lineages, which focus on basic cell structure and central metabolism, indicate an anaerobic, fermentative, saccharolytic lifestyle with the potential for degradation of (hemi)cellulose. This is consistent with the observed enrichment of OP9 on cellulosic biomass incubated in the hot spring GBS25 and supports the hypothesis that OP9 plays a role in thermophilic, cellulolytic microbial consortia. It is tempting to extend this prediction to other members of the OP9 lineage, which tend to be found in thermal, anaerobic environments with large amounts of biomass. The potential for utilization of xyloglucan may carve out a niche for OP9, and could explain its low-level but consistent presence in these environments. The apparent reliance of OP9 on exogenous vitamins suggests a dependence on other organisms in the in situ enrichments, corroborated by the presence of COGs involved in de novo vitamin synthesis elsewhere in the 77CS metagenome. These results facilitate enrichment strategies targeted at cultivation of OP9 and pave the way to understanding the role of this candidate phylum. This study also illustrates the utility of combining single-cell and metagenomics approaches, even in cases where datasets originate in distinct species or environments.
Based on data presented here, we propose the taxonomic epithets ‘Candidatus Caldatribacterium californiense’ and ‘Ca. Caldatribacterium saccharofermentans’ to refer to the OP9-cSCG and OP9-77CS phylotypes, respectively. The descriptions of the taxa are as follows: ‘Caldatribacterium’ (Cald.atri.bac.te’ri.um. L. adj. caldus, hot; L. adj. ater -tra -trum, black; L. neut. n. bacterium, rod or staff. N.L. neut. n. Caldatribacterium refers to a rod-shaped bacterium from a hot environment, where ‘black’ or ‘dark’ references both microbial ‘dark matter’ and the dark, anaerobic environments where the lineage is found), ‘saccharofermentans’ (sac.cha.ro. fer’men.tans. Gr. n. sakchâr, sugar; L. v. fermento, to ferment; N.L. part. adj. saccharofermentans, sugar-fermenting), and ‘californiense’ (ca.li.for.ni.en’se. N.L. neut. adj. californiense, of or belonging to California). 16S rRNA gene phylogenetics firmly places ‘Ca. Caldatribacterium saccharofermentans’ and ‘Ca. Caldatribacterium californiense’ within candidate bacterial phylum OP9 along with 16S rRNA gene phylotypes recovered from environments including geothermal systems, petroleum reservoirs, anaerobic digesters, and wastewater treatment facilities. In addition, phylogenetic analyses support the proposal for the candidate phylum ‘Atribacteria’ (A.tri.bac.te’ri.a. N.L. n. Atribacteria, the ‘dark’ bacterial phylum) inclusive of members of the OP9 lineage.
Methods
Single-cell sorting, whole genome amplification, and sequencing
An overview of methods used for a combined single-cell genomic and metagenomic analysis targeting OP9 is shown in Supplementary Figure S1. Samples for single-cell sorting were prepared from source pool sediment from LHC4 in the Long Valley Caldera, CA, USA19. Cells were separated from sediment by centrifugation over a Nycodenz density cushion45, stored either on ice/4 °C in ethanol (10%, volume/volume) or on dry ice/-80 °C in betaine (6%, weight/volume), and sorted within 1 week of collection. Measurements of temperature, pH, and conductivity of spring water at the collection site were performed using hand-held meters that were calibrated in the field prior to sampling (LaMotte 5 Series, Chestertown, MD or YSI Model 30, Yellow Springs, OH and WTW Model pH330i, Weilheim, Germany) as described previously18,19.
Single-cell sorting was performed on three separate weeks in October 2009, April 2010, and August 2011 using a microfluidic device mounted on a phase-contrast microscope equipped with a 1 W, 976 nm laser for optical trapping as previously described17, except that an updated but functionally analogous microfluidic device with 48 sorting chambers was used. Briefly, the cell sorting system involves loading a mixture of cells into each of two central channels, using the laser to transfer individual cells through gated channels into each of the 48 chambers, lysing them, and amplifying their genomes using MDA. Prior to loading into the microfluidic device, cells were diluted to ~104 cells/μL in PPT buffer (phosphate-buffered saline pH 7.4 with 0.01% pluronic F127 and 0.01% Tween-20) containing 0.1 mg/mL BSA (New England Biolabs, Ipswich, MA). In some cases, cells were pretreated with 0.5 μg/mL proteinase K to weaken proteinaceous cell envelopes and potentially enhance on-chip lysis46; this pretreatment did not apparently have a significant impact on lysis of cells containing OP9 16S rRNA genes. Additionally, cell samples sorted in April 2010 were mixed with 104 copies/μL of a traceable plasmid (pBK-CMV; Agilent Technologies, La Jolla, CA) to control for the presence of extra-cellular DNA. Cell lysis and whole genome amplification by MDA were performed in the microfluidic device as described17. MDA product subsamples (0.5 μL) were used as template in PCR to amplify either bacterial (primers 9bF and 1512uR) or archaeal (primers 8aF and 1406uR) 16S rRNA genes19. PCR products were sequenced from the forward primer using the Sanger method (Functional Biosciences, Madison, WI, USA) and compared to the NCBI non-redundant nucleotide (nr-nt) database using BLASTN47. Amplification products from April 2010 were additionally screened for the presence of the traceable plasmid containing a CMV promoter using primers 5′-CATATATGGAGTTCCGCGTTAC, 5′-CGTACTTGGCATATGATACACTTG, and a hydrolysis probe BHQ1-5′-CTGGCTGACCGCCCAACGA-HEX (vector reads were not detected in the MDA product mixtures from cells). Amplified SCGs belonging to the OP9 lineage were further amplified by a second round of amplification, and library preparation and shotgun sequencing was carried out as previously described17 using custom bar-coded adaptor oligonucleotides to enable pooling of multiple libraries and the 454 FLX platform with Titanium chemistry (Roche, Branford, CT) or alternatively, using the Nextera 454 titanium kit (Epicentre, Madison, WI). In all cases, purified libraries were quantified by digital PCR and normalized prior to sequencing48.
Assembly and co-assembly of single cell genomes
All pyrosequence reads from SCG libraries were initially filtered for quality using mothur49 and reads corresponding to trace contamination with human DNA were identified by BLASTN47 against the human genome database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/) and removed. Assembly of pyrosequence reads from individual SCGs was performed with the “Newbler” GS De Novo Assembler v2.6 (Roche), using default parameters with an expected coverage of 500 and the -urt option. FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to examine the %G+C distribution of reads, and the JCVI Multi-Dimensional Scatter Plot Viewer (http://gos.jcvi.org/openAccess/scatterPlotViewer.html)50 was used for TNF-PCA in assembled contigs. A subset of 10 SCGs with nearly identical %G+C read distributions and single, homogenous clustering of contigs by TNF-PCA, thus likely largely free of contamination, were selected. Contigs from these 10 SCGs were used to identify potential contaminants in four other SCGs using hierarchical average correlation clustering of TNF-PCA, with contigs cut into 2 kb fragments with 1 kb overlap. Reads mapping to contigs with one or more fragment falling outside the cluster overlapping with the 10 SCGs were removed, and filtered datasets were reassembled. Percent ANI of these 14 SCGs plus data from an additional, more recently sequenced OP9 SCG, were determined by pairwise BLASTN. The resulting distances (100% - %ANI) were used to construct a neighbor-joining tree using Phylip51 After further 454 sequencing on a subset of the 15 SCGs chosen for analysis, reads from these SCGs were pooled and a composite SCG (cSCG) assembly was made using Newbler. Additionally, a jackknifing procedure was used to remove chimeric reads from the cSCGs14.
Metagenome sequencing and assembly
Cellulosic biomass was incubated at various locations in the hot spring GBS, near Gerlach, NV, USA18. A full description of the cellulolytic enrichments is described in Peacock et al. (in review)25; methods pertaining to the metagenome used in this study are described below. 20 g of ammonia fiber explosion (AFEX)-treated corn stover (CS) sealed in a 100 micron mesh nylon filter bag was incubated buried ~1 cm in the sediment near the outflow of GBS for 64 days. Because the average water temperature was 77 °C at the site of incubation of the CS, this sample was designated “77CS”. After incubation, the 77CS substrate was harvested, frozen immediately on dry ice and stored at −80°C. DNA was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH), ethanol precipitated, and resuspended in TE buffer (10 mM Tris pH 8, 1 mM EDTA). 454 Rapid and Illumina standard libraries were prepared from purified DNA according to manufacturer’s protocols and sequenced on the 454 FLX platform (Roche) using Titanium chemistry and the Illumina GAIIx platform (Illumina, San Diego, CA) using paired-end 150 cycle reads, respectively. This produced a total of 179 Mb of 454 sequence and 13.1 Gb of Illumina data, which were co-assembled using a tiered assembly methodology. In brief, after trimming and quality filtering, the 454 and Illumina reads were initially assembled with Newbler and SOAPdenovo (http://soap.genomics.org.cn/soapdenovo.html), respectively. Contigs from the Newbler assembly and multiple SOAPdenovo assemblies using different k-mer lengths were dereplicated and merged using Newbler (contigs <2kb and remaining unassembled reads) followed by merging of all contigs with Minimus252. Thresholds for merging contigs was limited to overlaps of 60 bases or greater with >98% identity of the overlap. Validation of the assembly and estimation of contig coverage were performed using read mapping with Burrows-Wheeler Aligner and analyzed using Samtools53.
Metagenome binning and scaffolding of the composite single cell genome
TNF-PCA of the 77CS assembly with the same subset of 10 OP9 SCG contigs described above was performed. 77CS contigs falling within an ellipsoid with a centroid and semi-axis lengths defined by the mean and standard deviation of the first three principal components of the OP9 SCG contigs were binned as OP9-like, with the added constraint that 77CS contigs <2 kb were required to have a region of homology (>85% identity over >100 nucleotides) with the OP9-cSCG assembly. This subset of the metagenome, referred to as OP9-77CS, was compared to the OP9-cSCG by BLASTN to determine the reciprocal coverage and percent ANI. Further assembly and scaffolding of the cSCG contigs was performed manually using the output of the BLASTN analysis. In cases where cSCG contigs overlapped by comparison to the OP9-77CS contigs, the contigs were joined, using the region of the cSCG contig with a higher overall quality score in the region of overlap.
Single-cell genome and metagenome analysis
The OP9-cSCG and OP9-77CS contigs were submitted to RAST54 for gene calling and annotation. OP9-cSCG and OP9-77CS datasets were further filtered by BLASTP of predicted CDSs from one OP9 genome into a database containing the other OP9 genome, the RefSeq protein database (Release 52), and a recently sequenced Thiovulum single-cell genome14. Contigs where no CDSs had top BLASTP hits to the other OP9 genome, and where at least one CDS had a BLASTP hit with >70% identity to the RefSeq database or the Thiovulum genome, were removed. A single contig in the OP9-77CS containing a portion of a 23S rRNA gene with 98% identity to that of Thermodesulfobacterium sp. OPB45 was also removed; only 10 CDSs on 9 contigs in the OP9-77CS assembly had top BLASTP hits to Thermodesulfobacteria, all of which contained CDSs with top BLASTP hits to OP9-cSCG or other Bacteria, suggesting minimal contamination from this group in the OP9-77CS. Additional annotation of coding sequences predicted by RAST was performed using the KEGG Automatic Annotation Server (KAAS)55 using a BLAST bit score cutoff of 60, the Conserved Domain Database (CDD)56 with an e-value cutoff of 1e−5, and by comparison to a database of proteins assigned to Enzyme Commission (EC) numbers by Markov clustering using BLASTP with a 1e−20 cutoff57. A set of single copy genes conserved in Bacteria were detected and aligned using AMPHORA28, and phylogenies were inferred by maximum likelihood using RAxML V. 7.2.658. 16S rRNA genes from OP9 and other Bacteria were initially aligned using the Greengenes NAST server59. Distance matrices, bootstrapping, and maximum likelihood and neighbor-joining methods were performed using PHYLIP V. 3.6.951. A list of species and accession numbers of sequences used for phylogenetic analyses are shown in Supplementary Table S3. CDSs from OP9-cSCG and OP9-CS77 were analyzed by BLASTP against RefSeq protein database and the results were used to infer the phylogeny of the hits using CHNOSZ V. 0.9-760.
Supplementary Material
Acknowledgements
We thank David Mead (Lucigen Co.) and Bruce Dale (Michigan State University) for providing AFEX-treated corn stover; Joanna Tsai, Nicolas Gobet, Anastasia Nedderson for assistance with cell sorting and DNA sequencing; David and Sandy Jamieson for their generous access to GBS; the National Forest Service (Inyo National Forest, Mammoth Lakes Office) for permission to sample Little Hot Creek; and Jean Euzéby for advice on taxonomic nomenclature.
This research is supported by NASA Exobiology grant EXO-NNX11AR78G; U.S. National Science Foundation grants MCB 0546865 and OISE 0968421; U.S. Department of Energy (DOE) grant DE-EE-0000716; the Nevada Renewable Energy Consortium, funded by the DOE; and the Joint Genome Institute (CSP-182), supported by the Office of Science of the U.S. DOE under Contract No. DE-AC02-05CH11231. BPH was supported by a generous donation from Greg Fullmer. CAR was supported by National Institutes of Health Grant P20 RR-016464.
Footnotes
Author contributions J.A.D., P.C.B., S.R.Q. and B.P.H. conceived and designed the experiments. J.A.D. and B.P.H. collected and processed the samples. J.A.D. and P.C.B. sorted single cells and sequenced amplified DNA. J.A.D., P.C.B., S.K.M., and C.A.R. assembled and annotated single-cell genomes. S.G.T. and P.S.G.C. sequenced, assembled, and annotated metagenomes. S.K.M. and J.A.D. conducted phylogenomic and 16S rRNA gene phylogenetic analyses. S.K.M., J.A.D., and W.D.S. carried out bioinformatic predictions of cell structure. J.A.D. interpreted cell physiology. J.A.D. and B.P.H. wrote the paper with input and approval by all authors.
Accession numbers: 16S rRNA gene sequences of individual OP9 SCGs (Accession numbers KC110876 to KC110890) have been deposited in GenBank. Nearly-complete OP9 assemblies have been deposited as Whole Genome Shotgun projects at DDBJ/EMBL/GenBank under the accession numbers APCU00000000 (OP9-77CS) and APKF00000000 (OP9-cSCG). The versions described in this paper are the first versions, APCU01000000 (OP9-77CS) and APKF01000000 (OP9-cSCG). Chimera-filtered 454 reads used in the cSCG assembly have been deposited in the Short Read Archive (SRR609896). Annotated assemblies and predicted proteins for the OP9-cSCG (Genome ID 6666666.23228) and OP9-77CS (ID 6666666.23137) are available on the RAST guest account (http://rast.nmpdr.org, using login and password ‘guest’). Complete 77CS metagenome data are also available in the Integrated Microbial Genomes with Microbiome Samples (IMG/M, http://img.jgi.doe.gov/m) database, taxon object ID 3300000106.
Supplementary Information accompanies this paper at http://www.nature.com/naturecommunications (pending)
Competing financial interests: The authors declare no competing financial interests.
How to cite this article: (pending)
References
- 1.Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a yellowstone hot spring. J. Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Ann. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 3.Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 2008;6:431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 4.Marcy Y, et al. Dissecting biological ‘dark matter’ with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. PNAS. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lasken RS. Genomic sequencing of uncultured microorganisms from single cells. Nat. Rev. Microbiol. 2012;10:631–640. doi: 10.1038/nrmicro2857. [DOI] [PubMed] [Google Scholar]
- 6.Kalisky T, Blainey P, Quake SR. Genomic analysis at the single-cell level. Annu. Rev. Genet. 2011;45:431–445. doi: 10.1146/annurev-genet-102209-163607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hongoh Y, et al. Complete genome of the uncultured Termite Group 1 bacteria in a single host protist cell. PNAS. 2008;105:5555–5560. doi: 10.1073/pnas.0801389105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelletier E, et al. ‘Candidatus Cloacamonas acidaminovorans’: genome sequence reconstruction provides a first glimpse of a new bacterial division. J. Bacteriol. 2008;190:2572–2579. doi: 10.1128/JB.01248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takami H, et al. A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem. PLoS ONE. 2012;7:e30559. doi: 10.1371/journal.pone.0030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigue S, et al. Whole genome amplification and de novo assembly of single bacterial cells. PLoS ONE. 2009;4:e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blainey PC, Quake SR. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Res. 2011;39:e19–e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woyke T, et al. Decontamination of MDA reagents for single cell whole genome amplification. PLoS ONE. 2011;6:e26161. doi: 10.1371/journal.pone.0026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woyke T, et al. One bacterial cell, one complete genome. PLoS ONE. 2010;5:e10314. doi: 10.1371/journal.pone.0010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall IPG, Blainey PC, Spormann AM, Quake SR. A single-cell genome for. Thiovulum sp. Appl. Environ. Microbiol. 2012 doi: 10.1128/AEM.02314-12. doi:10.1128/AEM.02314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Podar M, et al. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl. Environ. Microbiol. 2007;73:3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegl A, et al. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. The ISME Journal. 2011;5:61–70. doi: 10.1038/ismej.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youssef NH, Blainey PC, Quake SR, Elshahed MS. Partial genome assembly for a candidate division OP11 single cell from an Anoxic spring (Zodletone Spring, Oklahoma) Appl. Environ. Microbiol. 2011;77:7804–7814. doi: 10.1128/AEM.06059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. doi: 10.1038/nature12352. in press. [DOI] [PubMed] [Google Scholar]
- 19.Vick TJ, Dodsworth JA, Costa KC, Shock EL, Hedlund BP. Microbiology and geochemistry of Little Hot Creek, a hot spring environment in the Long Valley Caldera. Geobiology. 2010;8:140–154. doi: 10.1111/j.1472-4669.2009.00228.x. [DOI] [PubMed] [Google Scholar]
- 20.Costa K, et al. Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles. 2009;13:447–459. doi: 10.1007/s00792-009-0230-x. [DOI] [PubMed] [Google Scholar]
- 21.Gittel A, Sørensen KB, Skovhus TL, Ingvorsen K, Schramm A. Prokaryotic community structure and sulfate reducer activity in water from high-temperature oil reservoirs with and without nitrate treatment. Appl. Environ. Microbiol. 2009;75:7086–7096. doi: 10.1128/AEM.01123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riviére D, et al. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. The ISME Journal. 2009;3:700–714. doi: 10.1038/ismej.2009.2. [DOI] [PubMed] [Google Scholar]
- 23.Tang Y-Q, et al. Characteristic microbial community of a dry thermophilic methanogenic digester: its long-term stability and change with feeding. Applied Microbiology and Biotechnology. 2011;91:1447–1461. doi: 10.1007/s00253-011-3479-9. [DOI] [PubMed] [Google Scholar]
- 24.Goris J, et al. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 25.Peacock JP, et al. Pyrosequencing reveals high-temperature cellulolytic microbial consortia in Great Boiling Spring after in situ lignocellulose enrichment. PLoS ONE. 2013;8:e59927. doi: 10.1371/journal.pone.0059927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatusov R, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu M, Eisen J. A simple, fast, and accurate method of phylogenomic inference. Genome Biology. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martín HG, et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- 30.Webster G, Parkes RJ, Fry JC, Weightman AJ. Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl. Environ. Microbiol. 2004;70:5708–5713. doi: 10.1128/AEM.70.9.5708-5713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutcliffe IC. Cell envelope architecture in the Chloroflexi: a shifting frontline in a phylogenetic turf war. Environ. Microbiol. 2011;13:279–282. doi: 10.1111/j.1462-2920.2010.02339.x. [DOI] [PubMed] [Google Scholar]
- 32.Bauer MW, et al. An endoglucanase, EglA, from the hyperthermophilic archaeon Pyrococcus furiosus hydrolyzes β-1,4 bonds in mixed-Linkage (1→3),(1→4)-β-d-glucans and cellulose. J. Bacteriol. 1999;181:284–290. doi: 10.1128/jb.181.1.284-290.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liebl W, et al. Analysis of a Thermotoga maritima DNA fragment encoding two similar thermostable cellulases, CelA and CelB, and characterization of the recombinant enzymes. Microbiology. 1996;142:2533–2542. doi: 10.1099/00221287-142-9-2533. [DOI] [PubMed] [Google Scholar]
- 34.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vlasenko E, Schülein M, Cherry J, Xu F. Substrate specificity of family 5, 6, 7, 9, 12, and 45 endoglucanases. Bioresource Technol. 2010;101:2405–2411. doi: 10.1016/j.biortech.2009.11.057. [DOI] [PubMed] [Google Scholar]
- 36.Moracci M, et al. Identification and molecular characterization of the first α-xylosidase from an archaeon. J. Biol. Chem. 2000;275:22082–22089. doi: 10.1074/jbc.M910392199. [DOI] [PubMed] [Google Scholar]
- 37.Erkens GB, Majsnerowska M, ter Beek J, Slotboom DJ. Energy coupling factor-type ABC transporters for vitamin uptake in prokaryotes. Biochemistry. 2012;51:4390–4396. doi: 10.1021/bi300504v. [DOI] [PubMed] [Google Scholar]
- 38.Vignais P. Hydrogenases and H2-reduction in primary energy conservation. Bioenergetics. 2008;45:223–252. doi: 10.1007/400_2006_027. [DOI] [PubMed] [Google Scholar]
- 39.Ma K, Weiss R, Adams MWW. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 2000;182:1864–1871. doi: 10.1128/jb.182.7.1864-1871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calusinska M, Happe T, Joris B, Wilmotte A. The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiology. 2010;156:1575–1588. doi: 10.1099/mic.0.032771-0. [DOI] [PubMed] [Google Scholar]
- 41.Schut GJ, Adams MWW. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 2009;191:4451–4457. doi: 10.1128/JB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biegel E, Schmidt S, González J, Müller V. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell. Mol. Life Sci. 2011;68:613–634. doi: 10.1007/s00018-010-0555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meier T, et al. Complete ion-coordination structure in the rotor ring of Na+-dependent F-ATP synthases. J. Mol. Biol. 2009;391:498–507. doi: 10.1016/j.jmb.2009.05.082. [DOI] [PubMed] [Google Scholar]
- 44.Luoto HH, Belogurov GA, Baykov AA, Lahti R, Malinen AM. Na+-translocating membrane pyrophosphatases are widespread in the microbial world and evolutionarily precede H+-translocating pyrophosphatases. J. Biol. Chem. 2011;286:21633–21642. doi: 10.1074/jbc.M111.244483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindahl V, Bakken LR. Evaluation of methods for extraction of bacteria from soil. FEMS Microbiology Ecology. 1995;16:135–142. [Google Scholar]
- 46.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White RA, 3rd, Blainey PC, Fan HC, Quake SR. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009;10:116. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inskeep WP, et al. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS ONE. 2010;5:e9773. doi: 10.1371/journal.pone.0009773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 5:164–166. [Google Scholar]
- 52.Sommer DD, Delcher AL, Salzberg SL, Pop M. Minimus: a fast, lightweight genome assembler. BMC Bioinformatics. 2007;8:64. doi: 10.1186/1471-2105-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aziz R, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Research. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swingley WD, et al. Coordinating environmental genomics and geochemistry reveals metabolic transitions in a hot spring ecosystem. PLoS ONE. 2012;7:e38108. doi: 10.1371/journal.pone.0038108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 59.DeSantis TZ, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dick JM. Calculation of the relative metastabilities of proteins using the CHNOSZ software package. Geochemical Transactions. 2008;9:10. doi: 10.1186/1467-4866-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.