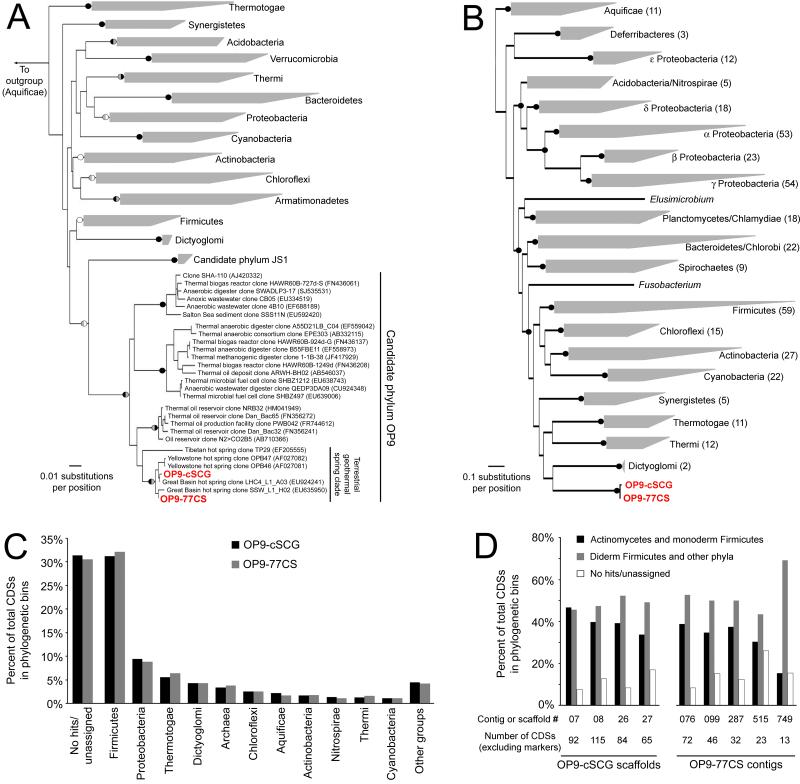
Figure 3.
Relationship of OP9-SCG and OP9-CS77 to other bacterial groups. (A) Neighbor-joining tree based on a distance matrix of 1349 aligned positions of 16S rRNA genes from selected bacterial phyla with cultured representatives and the candidate phyla JS1 and OP9, including those in the OP9-SCG and OP9-CS77 assemblies. Black (100%), grey (>80%), and white (>50%) circles indicate bootstrap support (100 pseudoreplicates) for selected nodes in phylogenies inferred using neighbor-joining (left half of circle) and maximum-likelihood (right half) methods. (B) Maximum-likelihood phylogeny inferred from concatenated alignments of predicted amino acid sequences of 31 housekeeping genes identified by AMPHROA in genomes representing a variety of bacterial phyla and the OP9-SCG and OP9-CS77 assemblies. The number of genomes represented in each wedge is indicated in parentheses, and black circles indicate bootstrap support of >80% for 100 pseudoreplicates. (C) Binning of CDSs in the OP9 assemblies based on the phylogenetic affiliation of their top BLASTP hit to a database of sequenced bacterial and archaeal genomes. Phylogenetic groups with fewer than 1% of top hits were aggregated (‘other groups’). (D) Phylogenetic binning of CDSs on contigs or scaffolds containing markers diagnostic for a diderm cell envelope structure31, emphasizing that top BLASTP hits were distributed among both monoderm and diderm Bacteria. Diderm Firmicutes includes members of the Negativicutes and Halanaerobiales.