Figure 1.
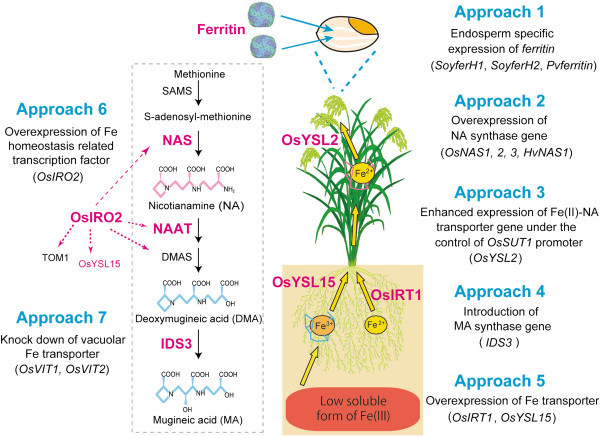
Seven transgenic approaches to Fe biofortification of rice. The pathway inside the gray dashed-line rectangle shows the biosynthetic pathway for mugineic acid family phytosiderophores (MAs) in graminaceous plants. SAMS, S-Adenosyl-methionine synthase; NAS, NA synthase; NAAT, NA aminotransferase; DMA, 2′-deoxymugineic acid; DMAS, DMA synthase; IDS3, MA synthase (dioxygenase that catalyzes the hydroxylation of DMA and epiHDMA at the 2′ position); Ferritin, iron storage protein; OsYSL2, Fe(II)-NA and Mn(II)-NA transporter; OsIRO2, Fe deficiency-inducible bHLH transcription factor related to Fe homeostasis in rice; OsIRT1, ferric transporter; OsYSL15, Fe(III)-DMA transporter; TOM1, MA transporter. Rice lacks the two dioxygenase genes (IDS2 and IDS3) and secretes only DMA. Approach 1: Enhancing Fe accumulation in seeds by introducing the Fe storage protein, ferritin gene, SoyferH1, SoyferH2 or Pvferritin, under the control of endosperm-specific promoters. Approach 2: Enhancing Fe transport within the plant body by the overexpression of NAS. Approach 3: Enhancing Fe influx to seeds by expression of the Fe(II)-NA transporter gene OsYSL2 under the control of the OsSUT1 promoter. Approach 4: Enhancing Fe uptake and translocation by introduction of the phytosiderophore synthase gene IDS3. Approach 5: Enhanced Fe uptake from soil by overexpression of the Fe transporter gene OsIRT1 or OsYSL15. Approach 6: Enhanced Fe uptake and translocation by overexpression of the OsIRO2 gene. Approach 7: Enhanced Fe translocation from flag leaves to seeds by knockdown of the vacuolar Fe transporter gene OsVIT1 or OsVIT2. The ferritin image was kindly provided by Dr. David S. Goodsell (Scripps Research Institute, La Jolla, CA, USA) and the RCSB PDB.
