Figure 1.
Dissecting Hematopoietic Differentiation
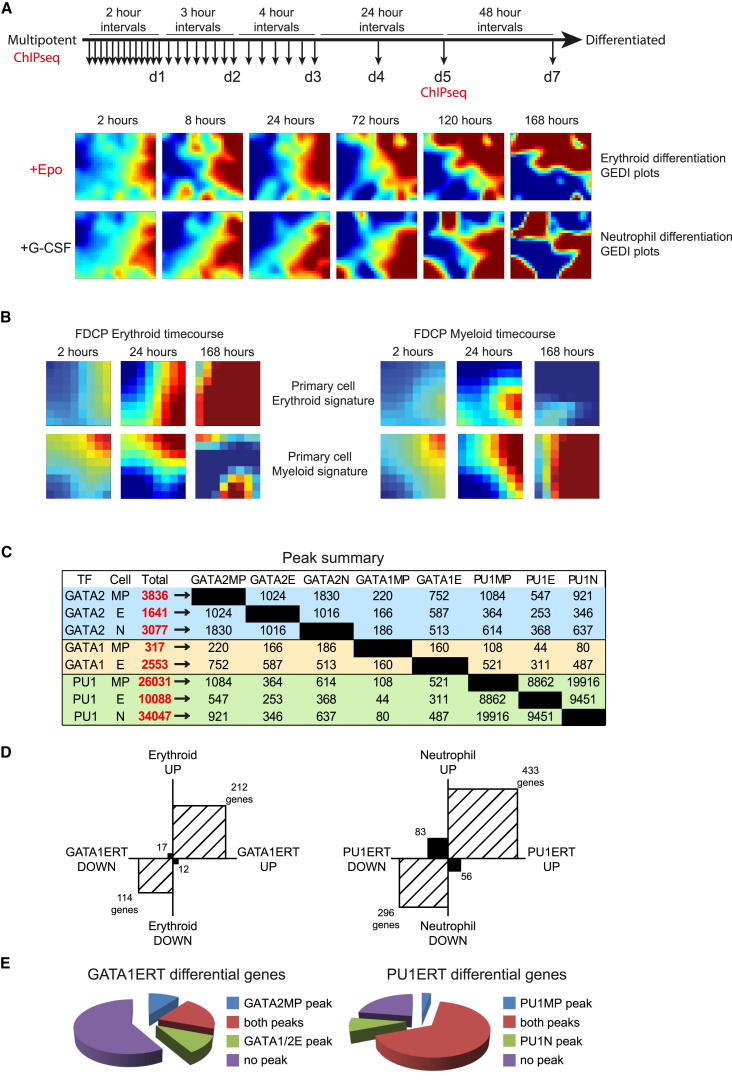
(A) Erythroid and neutrophil differentiation time course of FDCPmix cells. Time points for RNA (arrows) and ChIP-seq analysis are indicated. GEDI (Eichler et al., 2003) plots show changes in global transcriptomes through erythroid (upper) and neutrophil (lower) differentiation. Each pixel represents a group of coexpressed genes.
(B) Behavior of primary murine hematopoietic erythroid and myeloid signature genes in FDCPmix erythroid (left) and neutrophil (right) time courses.
(C) Peaks identified in each ChIP-seq experiment with their pairwise overlaps. MP, multipotent cells; E, erythroid cells (day 5); N, neutrophils (day 5).
(D) Genes modulated 2-fold by GATA1ERT induction in MP cells (left) were scored for up- or downregulation (d7/d0 >2 or <0.5) in the erythroid time course. The plot shows the number of genes with concordant (hatched) or discordant (black) regulation in the two experiments. PU.1ERT responses were compared against the neutrophil time course (right). See also Figure S2B.
(E) Left: GATA1ERT-responsive genes (as in D) with four GATA factor-binding profiles: bound only by GATA2 in MP cells (GATA2MP); bound by GATA2 in MP cells and GATA1 or GATA2 in E cells (both peaks); bound by GATA1 or GATA2 in E but not MP cells (GATA1/2E); and not bound (no peak). Right: PU.1ERT-responsive genes bound by PU.1 in MP cells (PU1MP), neutrophils (PU1N), or both.
See also Figures S1 and S2.