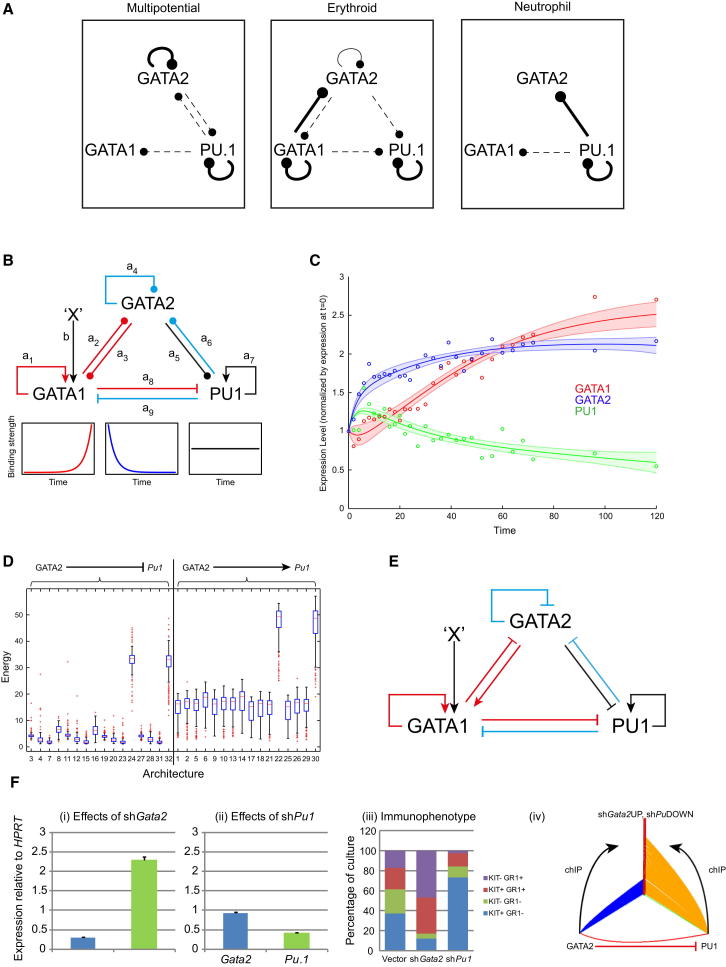
Figure 6.
Modeling the GATA1, GATA2, and PU.1 Triad
(A) Binding summary for GATA1, GATA2, and PU.1 over their own and each other’s loci, based on the ChIP-seq data. Bold, solid, and dotted connectors indicate strong, intermediate, and weak enrichments, respectively. See also Figures S6A and S6B.
(B) Basal architecture for the triad during erythroid differentiation based on binding data in (A) and the literature. Binding strengths (ax, b) were modeled as exponentially increasing (red), exponentially decreasing (blue), or constant (black), according to the changes observed between MP and E cells. Circled arrowheads, interactions of unknown sign based solely on DNA-binding data; bent arrows and blunt arrowheads, positive autoregulation and cross-inhibition of GATA1 and PU.1 as reported in the literature. X represents an undefined, but predicted, constant positive input to Gata1.
(C) Example of erythroid time course gene expression profile fits using the 60 best solutions for architecture 4 (see Table S6; Supplemental Experimental Procedures). Full lines, mean simulated expression; shaded contours, standard deviation; circles, experimental data points; red, Gata1; blue, Gata2; green, Pu.1.
(D) Energies for all 32 possible networks (Table S6), corresponding to the 200 parameter sets and grouped according to the sign of the GATA2-PU.1 interaction. Left: GATA2 represses Pu.1; right: GATA2 activates Pu.1. Whisker length is defined as 1.5× interquartile range.
(E) Example of a low-energy network (architecture 4) that provides a good fit (see C) to the observed expression data.
(F) Knockdown of Gata2 and Pu.1 in multipotent FDCPmix. Real-time quantitative RT-PCR analysis of Gata2 and Pu.1 expression following shRNA knockdown of (i) Gata2 or (ii) Pu.1. Expression normalized to Hprt and relative to the control vector, represented as mean ± SEM. (iii) Differentiation of MP cells assessed by surface antigen expression. shGata2 increased generation of Gr-1+ myeloid cells (either kit+ or kit−), whereas shPu.1 decreased myeloid output. (iv) Hive plot showing connectivity of GATA2 and PU.1 ChIP-seq to genes perturbed by shGata2/shPu.1 in multipotent FDCPmix. Red points (y axis) denote genes upregulated during neutrophil differentiation that are upregulated by shGata2 and downregulated by shPu.1. Blue and orange lines represent genes bound in MP cells by GATA2 and PU.1, respectively. The single red line represents upregulation of Pu.1 by shGata2.