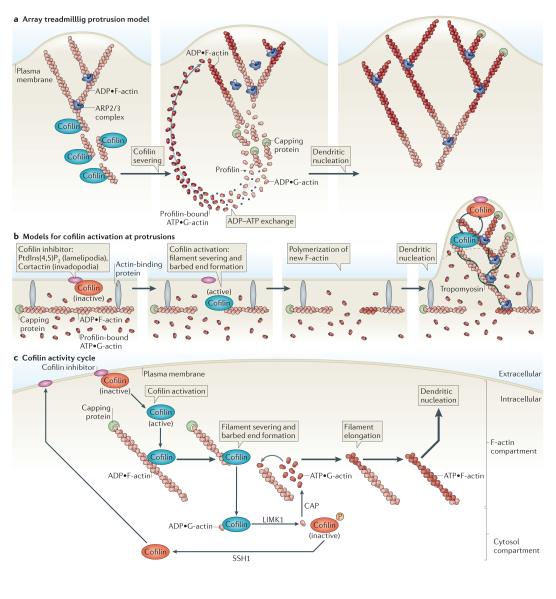
Figure 6. Models for cofilin function.
a ∣ Array treadmilling protrusion model. Cofilin severs and depolymerizes actin filaments at the base of the lamellipodium, thereby supplying G-actin monomers for steady-state actin polymerization in conditions of G-actin depletion. Dendritic nucleation is mediated by the actin-related protein 2/3 (ARP2/3) complex. b ∣ Models for cofilin activation at protrusions. Release of cofilin from its inhibitor (phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) at lamellipodia or cortactin at invadopodia) at the plasma membrane increases severing of actin filaments, generating free barbed ends that define the sites of dendritic nucleation by the ARP2/3 complex. G-actin monomers are supplied from an abundant pre-existing G-actin pool. The ARP2/3 complex mediates dendritic nucleation. Tropomyosin limits cofilin action, as it inhibits binding of cofilin to F-actin. This confines cofilin severing to the tip of protrusions (dotted black line), where cofilin continues its cycles of activation and deactivation due to the local cofilin activity cycle. c ∣ Cofilin activity cycle. Following its dephosphorylation or its release from inhibitory binding partners, activated cofilin binds to and severs actin filaments, generating barbed ends that are used for actin polymerization. LIM kinase 1 (LIMK1) and cyclase-associated protein (CAP) accelerate the dissociation of ADP•G-actin–cofilin complexes, and this increases the concentration of ATP•G-actin monomers available for filament elongation at the newly formed barbed ends. The capping of new barbed ends by actin interacting protein 1 (AIP1) (not shown) promotes cofilin-dependent filament disassembly. LIMK1 phosphorylates cofilin and thus inactivates it, leading to the release of bound G-actin. Upon the activation of slingshot 1 (SSH1) , cofilin is dephosphorylated and recycled to the inhibitory compartment. Figures are not drawn to scale. See BOX 1 for methods describing how to study cofilin activation.