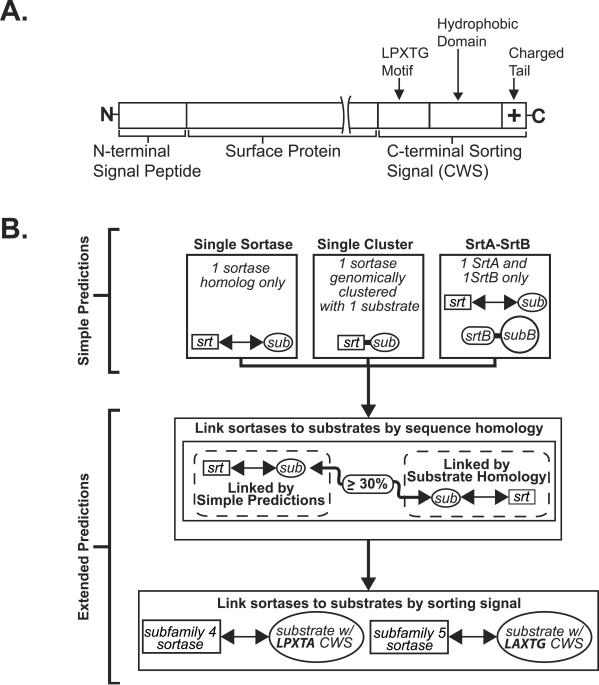
FIG. 1.
(A) Diagram illustrating a CWS-containing protein. It is composed of an N-terminal signal peptide and a C-terminal sorting signal that has a conserved LPXTG motif followed by a hydrophobic stretch of amino acids and positively charged residues at the C terminus. (B) A flowchart illustrating how functional linkages between sortase homologs and CWS-containing proteins were established. Three methods were used to make “simple” predictions: (i) single sortase, genome only contains a single sortase; (ii) single cluster, genome contains a gene cluster with a single sortase and a single CWS-containing protein; (iii) SrtA-SrtB, genome contains only SrtA and SrtB homologs. Two additional methods extended the initial predictions. With substrate sequence homology, a new functional link was made when a CWS-containing protein in one genome shared significant sequence homology (≥30%) to a previously assigned CWS-containing protein in another genome and both organisms encoded closely related sortases (see the text). With unique sorting signals, additional CWS-containing proteins were linked to their cognate sortase by examination of their sorting signal motif (subfamily-4 and subfamily-5 enzymes process the motifs LPXTA and LAXTG, respectively).